Leica TCS SP-5 X
High resolution confocal microscope Leica TCS SP5 equipped with four lasers (white light laser, UV lasers 355 nm and 405 nm, argon laser) represents unique equipment for studies related to chromatin biology. The important part of this system is a hood maintaining conditions necessary for cultivation of cell lines, thus enables to investigate processes in living cells. In our group we can perform various interesting experiments with such system:
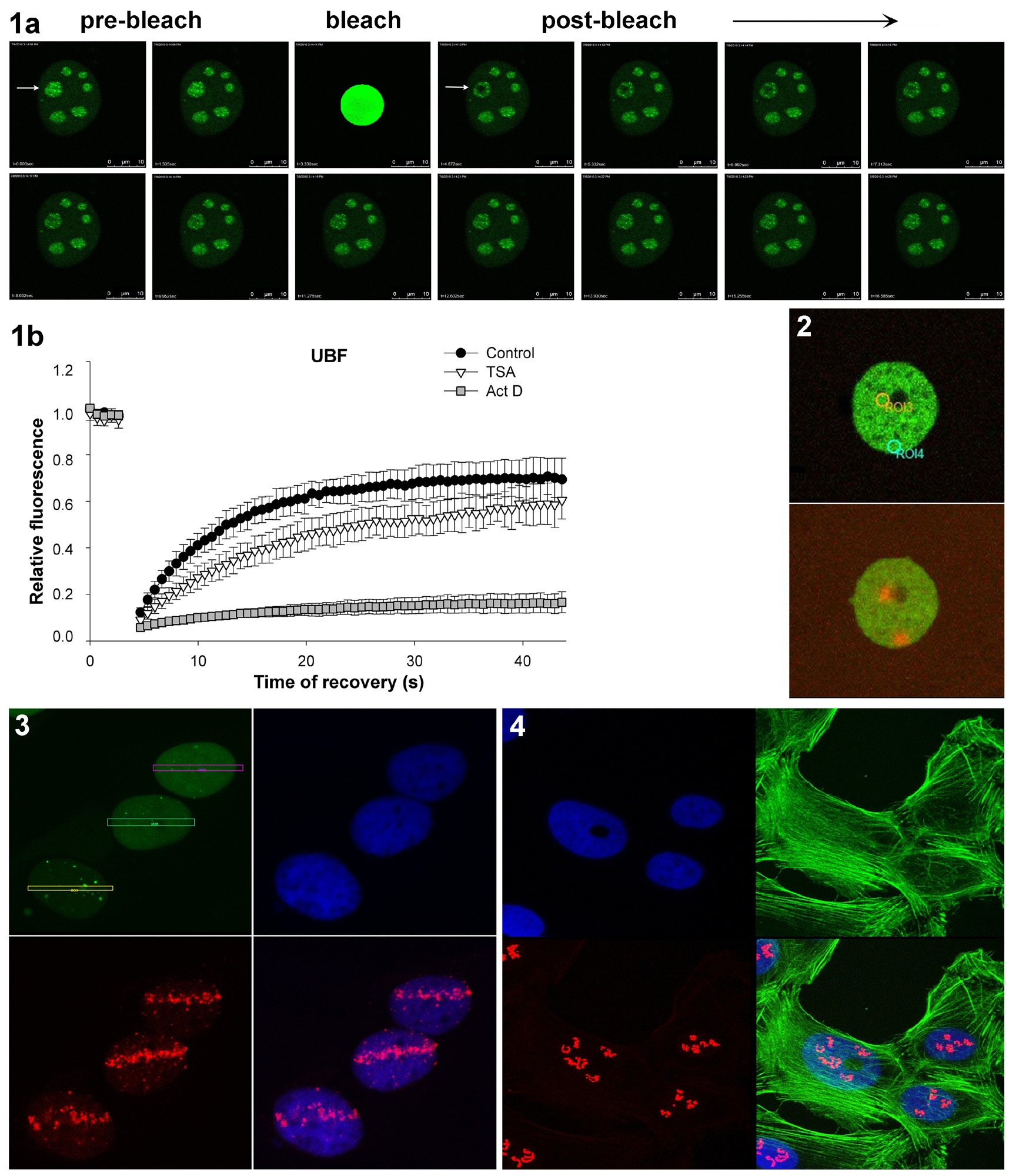
FRAP(Fluorescence Recovery After Photobleaching)
FRAP is used to measure the dynamics molecular mobility e.g. diffusion, transport or any other kind of movement of fluorescently labeled molecules in living cells. In this technique, the recovery of fluorescence in a defined region of interest (ROI) of a sample after a bleaching event is monitored by taking a time series of images. The recovery of fluorescence results from the movement of unbleached fluorophores from the surroundings into the bleached area. The speed of this mobility and the level of fully recovered intensity give information on mobile/immobile species of the fluorescent molecule. In our laboratory we use FRAP in combination with GFP-technology to investigate the kinetics properties of proteins in living cells (Fig. 1).Bleaching is performed with a 488-nm argon laser.
Photoconversion
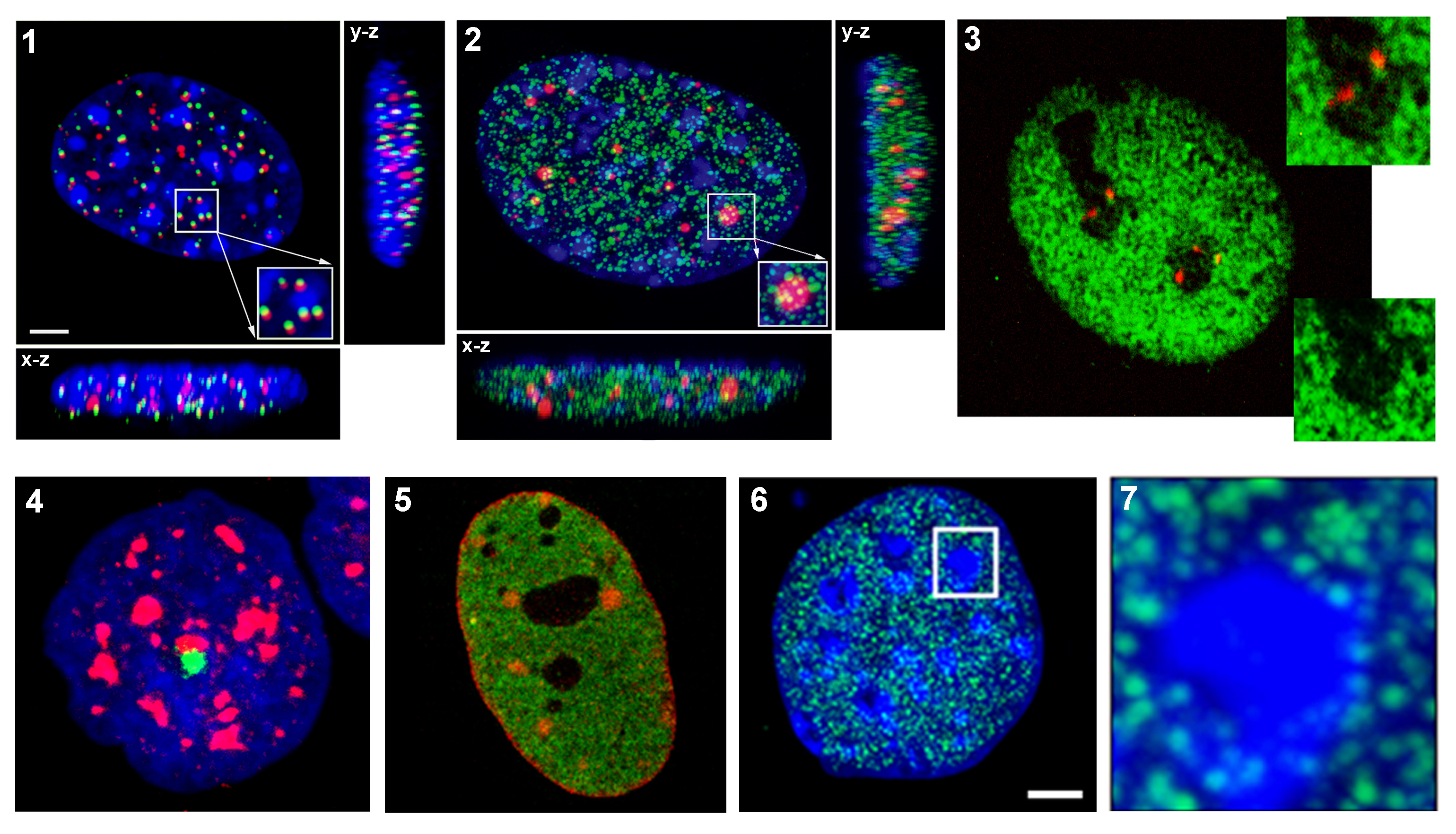
Photoconversion is the change of fluorescent features of fluorochromes after irradiation with UV laser. In our case we use Dendra2 fluorochrome which has excitation and emission maxima at 490 and 507 nm, analogous to EGFP or FITC. Dendra2 photoconversion appears after UV laser expozure of cells. Photoactivated Dendra2 protein is characteristic with excitation and emission maxima at 533 and 573 nm. Photoconversion experiments enable e.g. to track movement of cells in colony, or monitor changes in distribution of subcellular compartments and proteins in maternal and daughter cells, etc. In our experiments we monitor defined region of chromatin during the cell cycle and various treatments in living cells (Fig. 2).
Induction of double strand breaksin DNA(DSBs - Double Strand Breaks)
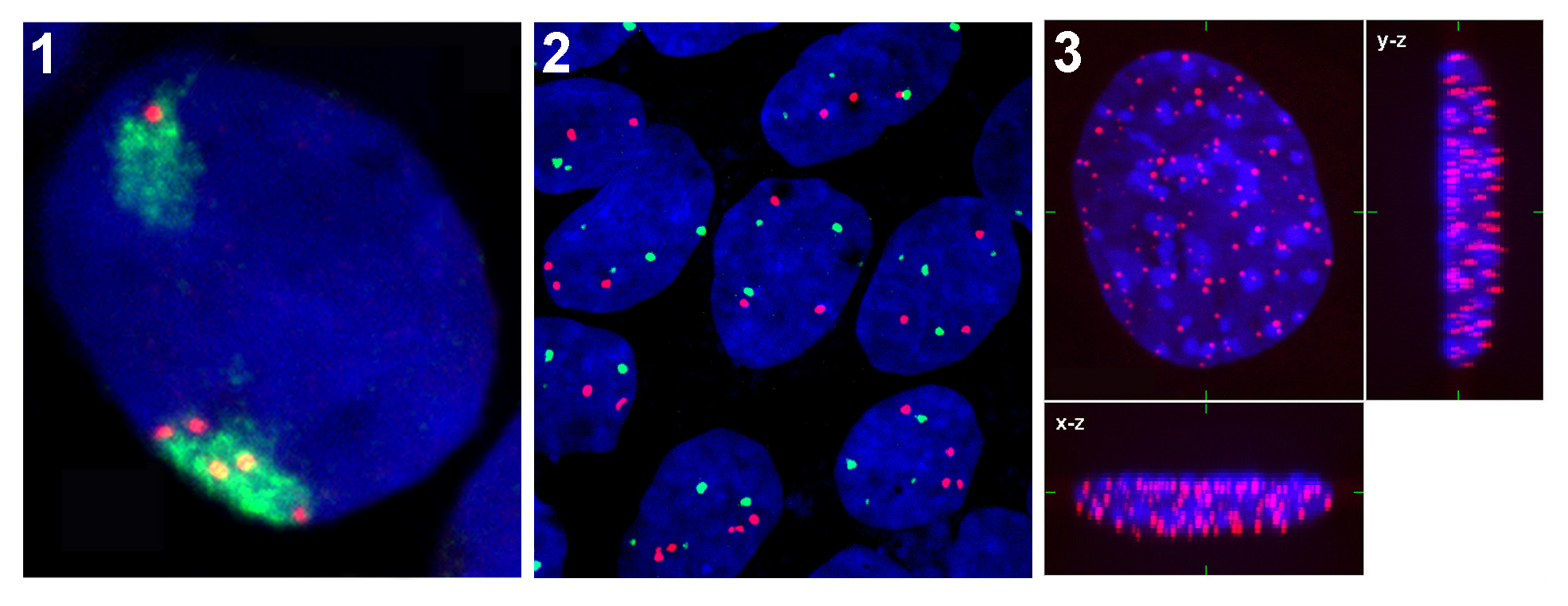
Most frequent causes of rise of double strand breaks are ionizing radiation and chemicals. These damages can lead to the cell death or mutations if they are not repaired. DNA repair processes are in long term the top topic of scientific interest and although many proteins have been identified to participate in these processes, a lot of functional aspects still remain elusive. Using UV laser (355 nm) can be induced double strand breaks in DNA and then it is possible to investigate DNA reparation processes on chromatin level directly in living cells (Fig. 3).
Immunocytochemistry
Immuno-fluorescence staining with specific antibodies is used for visualization of cell proteins. This method can be used also in combination with FISH or transfection technique (GFP fusion protein). In our laboratory we are focused mainly on study of nuclear proteins such as histone and heterochromatic proteins. Using high-resolution microscope Leica DMRXA we obtain high quality 3D images of nuclear structures and in dual-stained specimens we can evaluate a rate of colocalization of proteins.
FISH(Fluorescence In Situ Hybridization)
In our laboratory we use FISH technique for detection of various parts of the genome such as chromosomal territories, specific genes, or centromeric and telomeric areas of chromosomes. 3D Images are acquired with confocal system consisting of epifluorescent microscope Leica DMRXA with Nipkow disk-based confocal unit QLC100 and argon-krypton laser as a light source. Using special Acquiarium software we are able to analyze radial distributions of genetic elements in the cell nucleus (Matula et al. 2009).