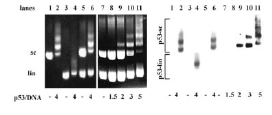
In the field of electrochemical analysis of nucleic acids DBCMO possesses the world primacy thanks to the discovery of DNA electroactivity, made by prof. Emil Paleček in the late 1950‘s [Naturwiss. 45 (1958), 186-187, Nature 188 (1960) 656-657]. This discovery showed that it was possible to observe signals for adenine, cytosine and guanine using oscillographic polarography at dropping mercury electrode. Since then, the department has been intensively dealing with the study of the behavior of nucleic acids at electrodes and in solutions which resulted in laying foundations of the field of nucleic acids electrochemistry.
After the initial period when only mercury electrode was used in DNA analysis, it was shown by Viktor Brabec and Glenn Dryhurst in the 1970s [Electroanal. Chem. 89 (1978), 161-173, Electroanal. Chem. 89 (1978), 161–173, Bioelectrochemistry Bioenerg. 8 (1981), 437–449] that DNA (bases adenine and guanine) yields analytically useful signals at carbon electrodes, too. Another development in the field of nucleic acids electrochemistry was achieved by introduction of adsorptive transfer stripping technique which led to reduced volume of the sample required for analysis and increased sensitivity [J. Electroanal. Chem. 214 (1986), 359–371].
Other areas of nucleic acids electrochemistry, in which the DBCMO contributed significantly, include:
- development of techniques for detection of DNA structure
- studies of electrochemical behavior of damaged DNA and development of sensors for detection of DNA damage
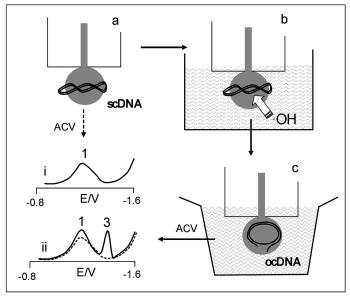
- development of electrochemical techniques for detection of DNA hybridization
- systematic studies of the peptide nucleic acid (PNA)
- labeling of nucleic acids with electrochemically active moieties. Osmium tetroxide complexes were the first labels used for electrochemical labeling of DNA, followed by a range of other DNA labels.
- introduction and application of "double-surface" electrochemical techniques (using magnetic beads) in studies of DNA hybridization and DNA-protein interactions.
- studies of electrochemical and interfacial behavior of synthetic oligonucleotides (ODN) with respect to nucleobase content and nucleotide sequence; studies of DNA, ODN and DNA components interactions with different electrode surfaces
The most recent contributions of the department to NA electrochemistry belong to these discoveries:
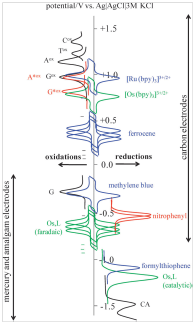
- carbon electrodes, namely pyrolytic graphite electrode in basal plane orientation, are suitable for studying not only anodic oxidation processes but also cathodic reduction of the nucleobases up to potential of -2.0 V [Electrochem. commun. 82 (2017), 34–38].
- DNA is capable of catalytic hydrogen evolution reaction, which can be observed as an electrocatalytic peak using voltammetry or chronopotentiometry at mercury and solid amalgam electrodes [ChemElectroChem 5 (2018), 936–942]
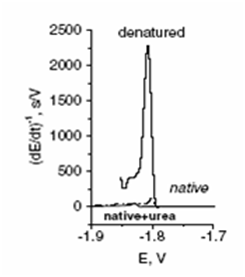
Another area of research with contributions from the DBCMO comprises the electrochemistry of proteins and novel methods for protein analysis. This was enabled by discovery of the so-called "peak H" [Electroanalysis 10 (1998), 403–409], which is yielded under certain conditions by peptides and proteins on mercury electrodes using constant current chronopotentiometric stripping analysis. This peak can be observed due to the ability of proteins and peptides to catalyse hydrogen evolution reaction. Peak H not only enables highly sensitive label-free detection of proteins, including those important in biomedicine and cancer research, but also specifically responses to structural changes of proteins, such as denaturation, aggregation, amino acid exchange, etc. This approach opened new possibilities of application of electrochemical methods for the study of protein-protein [Electrochim. Acta 297 (2019), 974–979] and/or protein-DNA interactions [Chem. Rev. 115 (2015), 2045–2108].
In the field of studies of the tumor suppressor protein p53, the DBCMO has a long tradition. In the 1990’s a new type of the interaction of p53 protein with DNA was discovered - the preferential p53 binding to supercoiled (sc) DNA [Oncogene 15 (1997), 2201–2209]. It has been found that p53 binds to scDNA not containing the specific binding site for p53 with much higher affinity than to linear DNA containing the same nucleotide sequence. Further studies have shown that the carboxy-terminal domain of the protein p53 and oligomeric state are crucial for this phenomenon and that DNA superhelicity affects sequence-specific binding of p53 to its target sites.