Electrochemistry and structure of nucleic acids
(Head of the group: Miroslav Fojta)
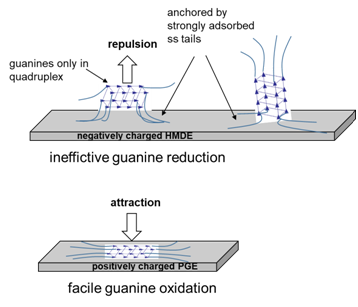
We are interested in relations between primary (sequence), secondary (global and local conformation) and/or tertiary (superhelicity, topology) DNA structure and its interactions with charged surfaces of electrodes. These interactions are reflected in electrochemical (oxidation, reduction) and/or surface (adsorption, 2D condensation) activities of DNA and its components. Up to recently, electrochemical reduction of nucleic acids was feasible at mercury-based electrodes while their oxidation was practically limited to application of carbon electrodes. Our recent findings have opened the possibility of studying nucleic acids reduction at pyrolytic graphite electrodes, making it possible to perform measurements of both reductions and oxidations at the same electrode material. On the other hand, now we can apply two different electrode materials in the cathodic region where particularly strong effects of DNA structure on the measured signals are observed, which extends the palette of variables to optimize conditions for structure-sensitive DNA analysis. We also pursue detailed studies of the effects of carbon electrode surface micro/nanostructure and surface chemistry on electrochemical responses of polymeric nucleic acids, their monomeric components and related groups of compounds.
Electrochemistry of modified nucleic acids
(Head of the group: Luděk Havran)
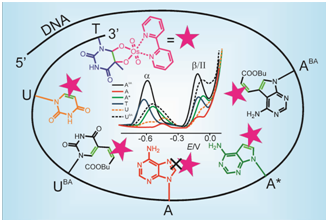
The group deals with application of chemically labeled DNA molecules for development of electrochemical biosensors. Labeled DNA is prepared by incorporation of chemically modified nucleobases using DNA polymerases or by direct modification of synthetic oligonucleotides. Incorporated modified nucleobases bear electrochemically active substituents or functional groups suitable for other modification reactions. This research also comprises studies of influence of chemical modification on DNA structure and its electrochemical behavior at different types of working electrodes. Development is focused mainly on the area of biochemical biosensors for monitoring of various DNA interactions (DNA hybridization, DNA damage, DNA-protein interactions).
Nanostructured surfaces for analysis of biomolecules
(Head of the group: Aleš Daňhel)
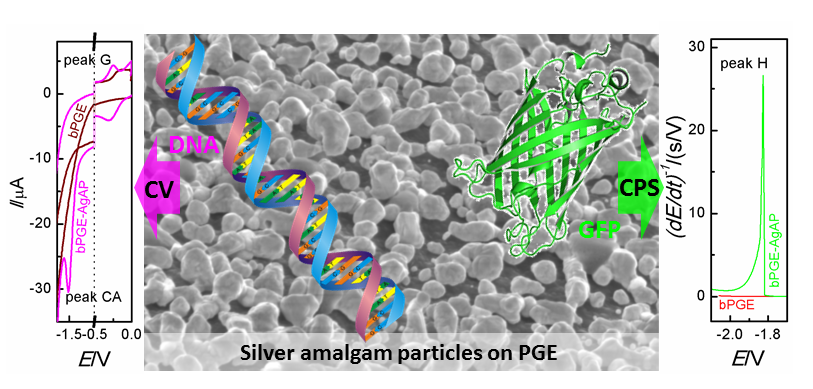
Research of the group is focused on studies of possibilities of preparation and properties of microcrystals and nanoparticles of silver amalgam deposited on chosen surfaces (various forms of carbon, metals (Ag, Au, Pt) and transparent materials (ITO, FTO)). The aim is to study and optimize conditions of electrochemical, chemical and vacuum steaming techniques for preparation of crystal microelectrodes and nanostructured surfaces with regard to their final morphology, chemical composition, electrochemical properties, spectral reflectivity and excitability of surface plasmons, which are crucial for their successful use in electrochemical and optical methods (spectroelectrochemistry, spectroscopic ellipsometry, surface plasmon resonance) for analysis of biomolecules as well as other species. Part of the project deals with studies of interactions of chosen organic compounds and biopolymers (DNA, RNA, proteins) with the surface of microcrystals and nanoparticles of silver amalgam in order to gain basic information important for potential development of novel electrochemical and optical (bio)sensors.
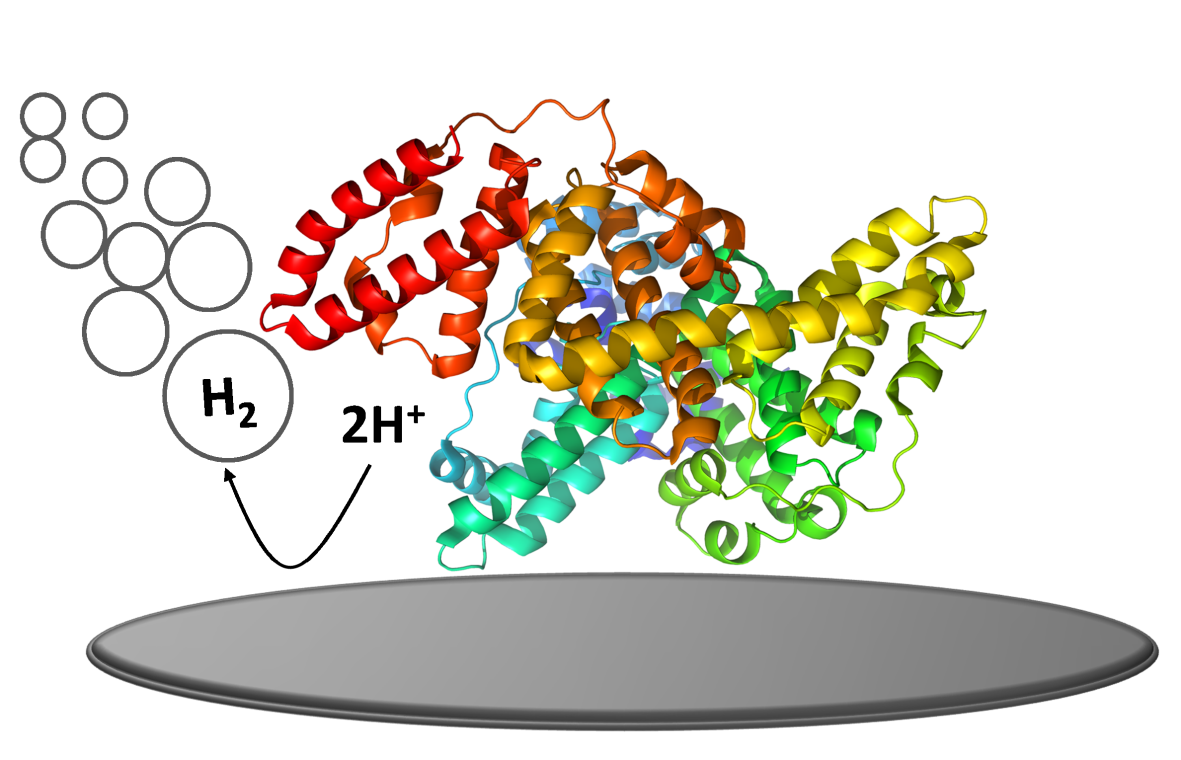
Catalytic hydrogen evolution of biomacromolecules and structure recognition of glycans in glycoprotein biomarkers
(Head of the group: Veronika Ostatná)
Influence of the structure of biomacromolecules (especially proteins and nucleic acids) and their complexes on catalytic hydrogen evolution at electrically charged surfaces. Discrimination of structure of glycans in glycoprotein cancer biomarkers. We also use label-based approaches, where proteins, predominantly glycoproteins and their glycan parts are labeled by osmium complexes.
Interaction of proteins with DNA
(Head of the group: Václav Brázda)
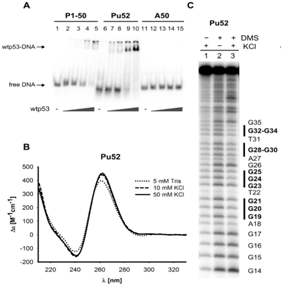
The main aims of the laboratory are to elucidate the binding preferences of various proteins to its target sequences and to local DNA structures, compare the DNA binding abilities of p53 isoforms to non-B DNA structures, study the stability and interaction partners interacting with local DNA structures with their influence to biological systems and development and validation of bioinformatics tools for analyses of nucleic acids and protein. Recently, we have published a tool for inverted repeats forming cruciforms and G4Hunter web for G-quadruplexes prediction. These tools are available at: bioinformatics.ibp.cz
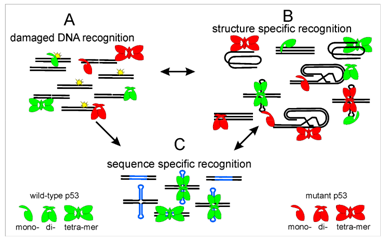
Brázda, V.; Coufal, J. Recognition of Local DNA Structures by p53 Protein. Int. J. Mol. Sci. 2017, 18, 375. https://www.ncbi.nlm.nih.gov/pubmed/28208646
Structure and topology of DNA in biology of p53 family and telomeres
(Head of the group: Marie Brázdová)
DNA structure and topology plays an important role in the biology of the p53 family and telomeres. The focus of the research group is to study the role of superhelicity and non-canonical structures of DNA and RNA, in particular G-quadruplexes and triplexes, in several areas. Above all, it is a study of their importance for tumor suppressor function of standard types of p53 family proteins. The second area is the analysis of these non-canonical structures as regulatory regions in the promoters of the target genes of the oncogenic mutant p53 proteins. In particular, we deal with the systematic study of the effect of G-quadruplex structure on its recognition not only by p53 proteins. The third area is the study of the role of p53, protein-protein interactions and G-quadruplexes in telomeric biology.