Research profile
The Department of Biophysics of Nucleic Acids studies structural, thermodynamic and interaction properties of deoxyribonucleic (DNA) and ribonucleic acid (RNA). DNA serves as a carrier of genetic information of the organisms, whereas RNA holds broad variety of functions depending on RNA type. DNA can exist in conformations (secondary structures) substantially different from the classical double helix, reported in 1953 by James Watson and Francis Crick. These conformations differ in type of base pairing helix, number of associated strands and their mutual orientation, structure handedness and others. Some of these conformations, reported for DNA, were observed also for RNA. The capability of particular nucleotide primary sequences to adopt specific conformations is critical for their recognition by other molecules, especially proteins and low-molecular weight drugs. Such unusual conformations have been shown to be critical in both normal and pathological phenomena, including replication, transcription, differentiation, aging, various types of cancer and other severe diseases.
The conformation of nucleic acids is strongly reflected by spectroscopic methods in the ultra violet light region, particularly by spectroscopy of circular dichroism (CD). CD originates from different absorption of left-handed and right-handed circularly polarized light by chiral molecules. Dominant part of CD comes from the chirality linked to asymmetric stacking of absorbing bases into different helical arrangements and only minor part of comes from the chiral sugar carbon atom to which the heterocyclic base attaches in nucleosides. As a result, each conformation of nucleic acids gives specific CD spectrum. The CD spectroscopy itself, however, cannot fully follow all the aspects of the conformations and we use a set of complementary methods to bring more detailed description of the unusual DNA / RNA conformations.
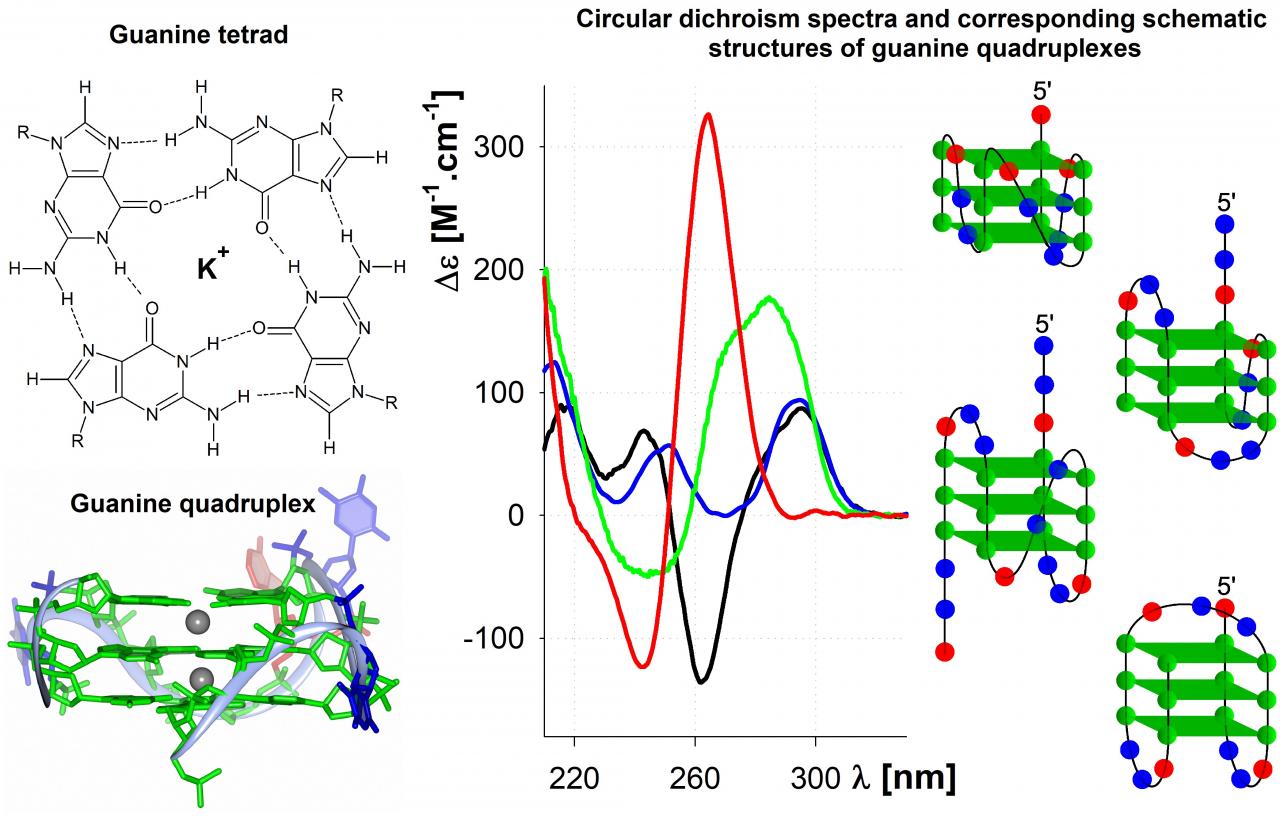
Recently we give most focus to the guanine quadruplexes. Guanine quadruplexes are secondary structures of nucleic acids, composed of several stacked square-shaped guanine tetrads, each formed from four Hoogsteen hydrogen bonded guanines. Whole structure is stabilized by some cations, especially potassium one, as well as by crowding conditions. These conditions facilitate the potential quadruplex occurrence in vivo. The potential quadruplex forming sequences (PQS) were found in genomes of numerous species with high occurrence at telomeres and gene promoters. Consequently, the guanine quadruplexes were indirectly observed both in purified genomic DNA and in cells. Based on the particular sequence, various types of quadruplexes were described, differing in detailed structure, as well as, in biophysical and biochemical properties.
During our long-lasting studies of quadruplexes formed by telomere sequences, we have shown that the conformation of human telomere quadruplex strongly depends on DNA concentration used, which clarified the differences in reports using various methods. Consecutively, we determined the effect of various naturally occurring base lesions on the structure of the telomere quadruplex, as well as we verified other mechanism, by which a repetitive sequence like telomere might handle base lesion. Recently, we described an inhibitory effect of guanine lesions on a molecular crowding-induced conformational switch of guanine quadruplexes. Such effect might have serious effect on potential interactions of the quadruplex. On a different system, we have shown that guanine-rich region present at the transcription-start site of the oct4 gene promoter forms in vitro parallel guanine quadruplex of substantial thermodynamic stability even in context of double-helical surrounding. The quadruplex serve as a positive regulator of expression of the oct4 gene and its stability and function might be modulated by small-molecule ligand. Recently, we started a project aiming on the identification of guanine quadruplexes and their role in the genome of the tick-borne encephalitis virus.
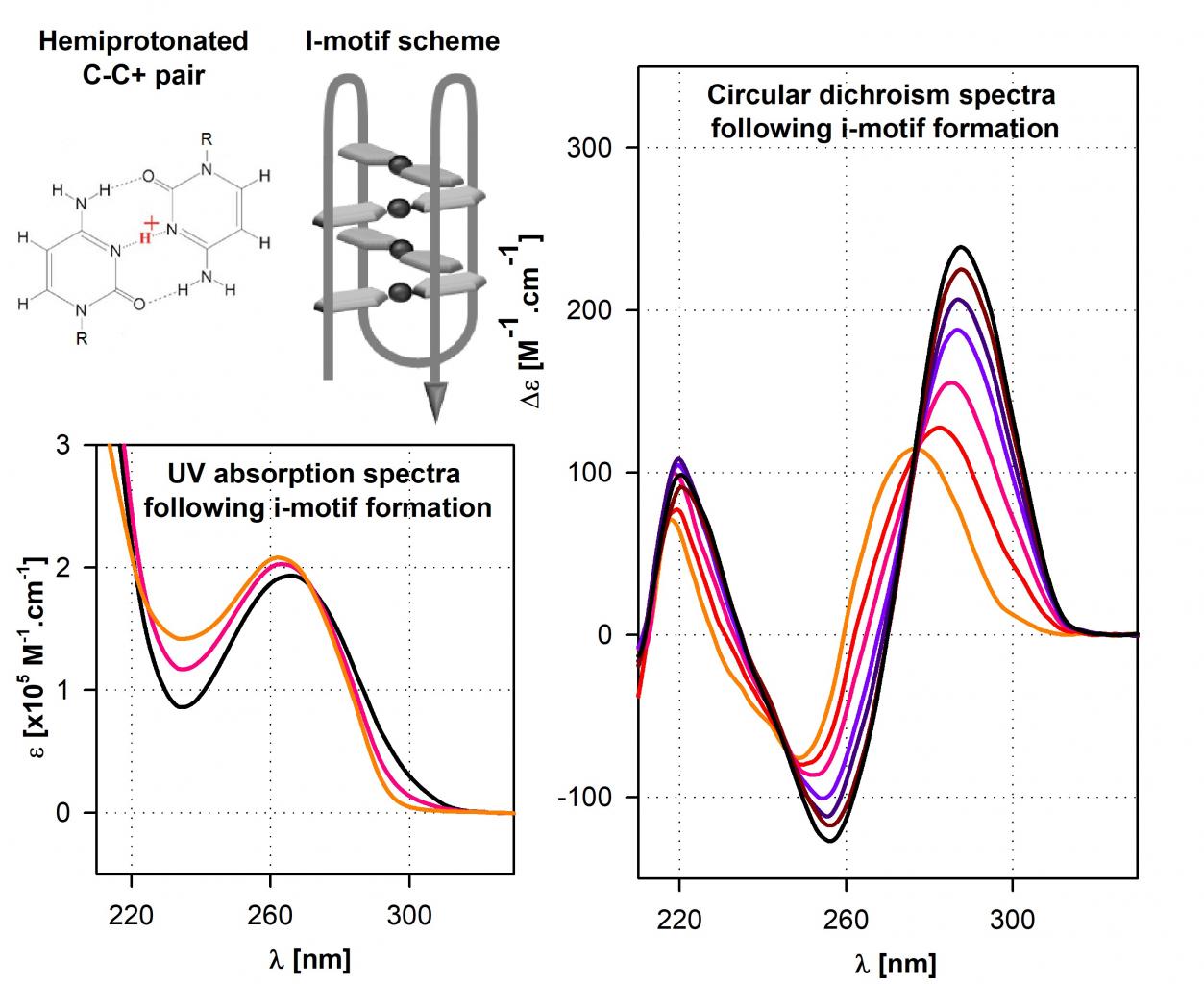
In context of double stranded nucleic acid, when one strand is guanine rich and prone to form guanine quadruplex, the complementary strand is cytosine rich. Such C-rich strands are known for more than 20 years to form unusual secondary structure called cytosine i-motif. I-motifs are based on a pairing of non-protonated and protonated cytosine, i.e. hemiprotonated cytosine pairs, that are stacked over each other in an intercalated way. Cytosine i-motifs were recently observed in fixed cells using antibodies. We recently reported exhaustive rules of relationship between primary sequence and thermodynamic stability of DNA i-motifs, together with an unusual multi-state behaviour of i-motifs formed by sequences with long stretches of cytosine. In addition, we reported the influence of various naturally occurring base lesions on the stability of i-motif formed by human telomere sequence.
One of the main tasks in the research filed is to confirm the existence of these unusual structures of nucleic acids directly in cells, if possible, at nucleoside resolution. Several approaches were reported, using structure-selective small molecules, enzymes or antibodies. We are currently implementing some of these approaches in our laboratory on DNA or RNA genomes.
Unusual secondary structures in genome or transcriptome represent also potential targets of cellular proteins. Many of them were already described, including helicases, transcription factors or structural proteins. In case of the latter two examples, potentially acting through interaction with guanine quadruplexes, most of them share one of two reported structural motifs: zinc-finger motif or RGG domains. In our research, we study two examples of such proteins, one of each group, to put more light on the protein interaction with guanine quadruplexes in context of their conformation.
Historically, the laboratory has been interested in the conformations adopted by simple, but biologically relevant, sequence repeats. For example, expansion of certain trinucleotide repeats correlates with numerous human diseases. These pathological expansions include the (CAG)n/(CTG)n sequences linked to Huntington’s and Kennedy’s diseases, myotonic dystrophy, spinocerebellar ataxia and others; the (CCG)n/(CGG)n sequences connected with fragile X syndrome; and the (AAG)n/(CTT)n sequences that play a role in Friedreich’s ataxia. We continue our research in these short repeats by novel approaches.
