Cluster no. 4
Go back to cluster table
Cluster is part of supercluster: 3
Cluster characteristics:
| size |
11887 |
| size_real |
11887 |
| ecount |
353544 |
| supercluster |
3 |
| annotations_summary |
8.30% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-INT
5.61% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-CHDII
0.04% organelle/plastid
0.01% Class_I/LTR/Ty1_copia/SIRE:Ty1-RH
0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RH
0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-INT
0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RT
|
| pair_completeness |
0.576316138443177 |
| pbs_score |
None |
| TR_score |
None |
| TR_monomer_length |
None |
| loop_index |
0.00471102885505174 |
| satellite_probability |
1.00051650786948e-21 |
| consensus |
None |
| TAREAN_annotation |
Other |
| orientation_score |
1 |
comparative analysis:

Comparative analysis - species read counts:
| Species |
Read count |
| S8 |
3920 |
| S5 |
21 |
| S4 |
17 |
| S7 |
3820 |
| S9 |
4060 |
| S6 |
20 |
| S2 |
20 |
| S1 |
6 |
| S3 |
4 |
comparative analysis - number of edges between species:
|
S8 |
S5 |
S4 |
S7 |
S9 |
S6 |
S2 |
S1 |
S3 |
| S8 |
38700 |
184.0 |
116.0 |
37200.0 |
39700 |
259.0 |
163.0 |
44.0 |
15.0 |
| S5 |
184 |
1.0 |
1.0 |
217.0 |
212 |
2.0 |
0.5 |
0.0 |
0.0 |
| S4 |
116 |
1.0 |
0.0 |
118.0 |
110 |
1.0 |
1.5 |
1.0 |
0.0 |
| S7 |
37200 |
217.0 |
118.0 |
36600.0 |
38900 |
240.0 |
148.0 |
37.5 |
18.5 |
| S9 |
39700 |
212.0 |
110.0 |
38900.0 |
42000 |
254.0 |
147.0 |
40.0 |
20.0 |
| S6 |
259 |
2.0 |
1.0 |
240.0 |
254 |
1.0 |
0.0 |
0.5 |
0.0 |
| S2 |
163 |
0.5 |
1.5 |
148.0 |
147 |
0.0 |
1.0 |
0.0 |
0.0 |
| S1 |
44 |
0.0 |
1.0 |
37.5 |
40 |
0.5 |
0.0 |
1.0 |
0.0 |
| S3 |
15 |
0.0 |
0.0 |
18.5 |
20 |
0.0 |
0.0 |
0.0 |
0.0 |
comparative analysis - observed/expected number of edges between species
|
S8 |
S5 |
S4 |
S7 |
S9 |
S6 |
S2 |
S1 |
S3 |
| S8 |
1.010 |
0.905 |
1.010 |
0.996 |
0.994 |
1.040 |
1.070 |
1.080 |
0.852 |
| S5 |
0.905 |
0.927 |
1.650 |
1.100 |
1.000 |
1.510 |
0.621 |
0.000 |
0.000 |
| S4 |
1.010 |
1.650 |
0.000 |
1.060 |
0.921 |
1.340 |
3.310 |
8.190 |
0.000 |
| S7 |
0.996 |
1.100 |
1.060 |
1.010 |
0.998 |
0.990 |
1.000 |
0.943 |
1.080 |
| S9 |
0.994 |
1.000 |
0.921 |
0.998 |
1.010 |
0.975 |
0.929 |
0.940 |
1.090 |
| S6 |
1.040 |
1.510 |
1.340 |
0.990 |
0.975 |
0.616 |
0.000 |
1.880 |
0.000 |
| S2 |
1.070 |
0.621 |
3.310 |
1.000 |
0.929 |
0.000 |
1.660 |
0.000 |
0.000 |
| S1 |
1.080 |
0.000 |
8.190 |
0.943 |
0.940 |
1.880 |
0.000 |
23.000 |
0.000 |
| S3 |
0.852 |
0.000 |
0.000 |
1.080 |
1.090 |
0.000 |
0.000 |
0.000 |
0.000 |
protein domains:
protein domains:

Reads annotation summary
| |
cl_string |
domain |
Freq |
proportion |
| plastid
|
plastid
|
|
5
|
4.2e-04
|
| SIRE Ty1-RH
|
SIRE
|
Ty1-RH
|
1
|
8.4e-05
|
| Tekay Ty3-CHDII
|
Tekay
|
Ty3-CHDII
|
667
|
5.6e-02
|
| Reina Ty3-INT
|
Reina
|
Ty3-INT
|
1
|
8.4e-05
|
| Tekay Ty3-INT
|
Tekay
|
Ty3-INT
|
987
|
8.3e-02
|
| Tekay Ty3-RH
|
Tekay
|
Ty3-RH
|
1
|
8.4e-05
|
| Tekay Ty3-RT
|
Tekay
|
Ty3-RT
|
1
|
8.4e-05
|
|
clusters with similarity:
| Cluster |
Number of similarity hits |
|
| 32 |
796 |
| 40 |
582 |
| 167 |
286 |
| 79 |
261 |
| 166 |
148 |
| 104 |
141 |
| 9 |
100 |
| 53 |
95 |
| 114 |
66 |
| 21 |
57 |
| 221 |
40 |
| 103 |
29 |
| 247 |
23 |
| 203 |
21 |
| 287 |
10 |
| 12 |
4 |
| 209 |
3 |
| 387 |
2 |
| 51 |
1 |
| 213 |
1 |
|
clusters connected through mates:
| Cluster |
Number of shared
read pairs |
k |
|
| 40 |
343 |
0.158 |
| 32 |
257 |
0.117 |
| 167 |
144 |
0.0798 |
| 114 |
140 |
0.072 |
| 79 |
134 |
0.074 |
| 104 |
116 |
0.065 |
| 166 |
108 |
0.0611 |
| 103 |
92 |
0.0497 |
| 203 |
86 |
0.0489 |
| 9 |
82 |
0.0357 |
| 53 |
78 |
0.0362 |
| 221 |
52 |
0.0305 |
| 209 |
43 |
0.0249 |
| 12 |
36 |
0.0129 |
| 213 |
36 |
0.0206 |
| 75 |
25 |
0.0113 |
| 11 |
22 |
0.00775 |
| 287 |
19 |
0.0115 |
| 5 |
17 |
0.00492 |
| 247 |
16 |
0.00976 |
| 8 |
15 |
0.00439 |
| 22 |
12 |
0.00372 |
| 17 |
9 |
0.0028 |
| 36 |
9 |
0.00355 |
| 78 |
9 |
0.00335 |
| 18 |
8 |
0.0028 |
| 157 |
8 |
0.00398 |
| 21 |
7 |
0.00337 |
| 59 |
7 |
0.00253 |
| 68 |
7 |
0.00293 |
| 102 |
6 |
0.00305 |
| 106 |
6 |
0.003 |
| 473 |
6 |
0.00373 |
| 1730 |
6 |
0.00374 |
| 2470 |
6 |
0.00375 |
| 54 |
5 |
0.00206 |
| 65 |
5 |
0.0025 |
| 139 |
5 |
0.00269 |
| 3540 |
5 |
0.00312 |
| 69 |
4 |
0.00147 |
| 91 |
4 |
0.00178 |
| 100 |
4 |
0.00197 |
| 140 |
4 |
0.00196 |
| 172 |
4 |
0.00241 |
| 2130 |
4 |
0.0025 |
| 3040 |
4 |
0.0025 |
| 4500 |
4 |
0.0025 |
| 5880 |
4 |
0.0025 |
| 51 |
3 |
0.00107 |
| 83 |
3 |
0.00152 |
| 150 |
3 |
0.00172 |
| 199 |
3 |
0.00177 |
| 205 |
3 |
0.00181 |
| 387 |
3 |
0.00186 |
| 1740 |
3 |
0.00187 |
| 4670 |
3 |
0.00187 |
| 10000 |
3 |
0.00188 |
| 11200 |
3 |
0.00188 |
| 17700 |
3 |
0.00188 |
| 162 |
2 |
0.000953 |
| 173 |
2 |
0.00111 |
| 182 |
2 |
0.00113 |
| 192 |
2 |
0.00113 |
| 206 |
2 |
0.00112 |
| 261 |
2 |
0.00122 |
| 379 |
2 |
0.00123 |
| 395 |
2 |
0.00124 |
| 1750 |
2 |
0.00125 |
| 2530 |
2 |
0.00125 |
| 6080 |
2 |
0.00125 |
| 9530 |
2 |
0.00125 |
| 10000 |
2 |
0.00125 |
| 10500 |
2 |
0.00125 |
| 12700 |
2 |
0.00125 |
| 13300 |
2 |
0.00125 |
| 13600 |
2 |
0.00125 |
| 15700 |
2 |
0.00125 |
| 17500 |
2 |
0.00125 |
| 17700 |
2 |
0.00125 |
| 18200 |
2 |
0.00125 |
| 19800 |
2 |
0.00125 |
| 21300 |
2 |
0.00125 |
| 24400 |
2 |
0.00125 |
| 34700 |
2 |
0.00125 |
| 38400 |
2 |
0.00125 |
| 50000 |
2 |
0.00125 |
| 57000 |
2 |
0.00125 |
| 57300 |
2 |
0.00125 |
| 63700 |
2 |
0.00125 |
| 67100 |
2 |
0.00125 |
| 30 |
1 |
0.000384 |
| 44 |
1 |
0.000408 |
| 46 |
1 |
0.000399 |
| 107 |
1 |
0.000461 |
| 138 |
1 |
0.000555 |
| 198 |
1 |
0.000562 |
| 235 |
1 |
0.000595 |
| 246 |
1 |
0.000592 |
| 253 |
1 |
0.000603 |
| 256 |
1 |
0.000599 |
| 262 |
1 |
0.000609 |
| 280 |
1 |
0.000601 |
| 381 |
1 |
0.000618 |
| 415 |
1 |
0.000615 |
| 439 |
1 |
0.00062 |
| 472 |
1 |
0.000621 |
| 497 |
1 |
0.000621 |
| 519 |
1 |
0.000617 |
| 546 |
1 |
0.00062 |
| 549 |
1 |
0.000621 |
| 554 |
1 |
0.000621 |
| 636 |
1 |
0.00062 |
| 710 |
1 |
0.000623 |
| 800 |
1 |
0.000622 |
| 823 |
1 |
0.000622 |
| 966 |
1 |
0.000623 |
| 1260 |
1 |
0.000623 |
| 1350 |
1 |
0.000624 |
| 1450 |
1 |
0.000623 |
| 1490 |
1 |
0.000625 |
| 1510 |
1 |
0.000624 |
| 1520 |
1 |
0.000624 |
| 1590 |
1 |
0.000624 |
| 1700 |
1 |
0.000624 |
| 2040 |
1 |
0.000624 |
| 2170 |
1 |
0.000624 |
| 2230 |
1 |
0.000625 |
| 2400 |
1 |
0.000624 |
| 2450 |
1 |
0.000624 |
| 2670 |
1 |
0.000625 |
| 2900 |
1 |
0.000625 |
| 3330 |
1 |
0.000624 |
| 3640 |
1 |
0.000625 |
| 3650 |
1 |
0.000625 |
| 3860 |
1 |
0.000625 |
| 4050 |
1 |
0.000625 |
| 4160 |
1 |
0.000625 |
| 4290 |
1 |
0.000625 |
| 4840 |
1 |
0.000625 |
| 4940 |
1 |
0.000625 |
| 5010 |
1 |
0.000625 |
| 5380 |
1 |
0.000625 |
| 5510 |
1 |
0.000625 |
| 5660 |
1 |
0.000625 |
| 5670 |
1 |
0.000625 |
| 6000 |
1 |
0.000625 |
| 6030 |
1 |
0.000625 |
| 6320 |
1 |
0.000625 |
| 6430 |
1 |
0.000625 |
| 6500 |
1 |
0.000625 |
| 6590 |
1 |
0.000625 |
| 7160 |
1 |
0.000625 |
| 8140 |
1 |
0.000625 |
| 8340 |
1 |
0.000625 |
| 8440 |
1 |
0.000625 |
| 9110 |
1 |
0.000625 |
| 9370 |
1 |
0.000626 |
| 10100 |
1 |
0.000625 |
| 10600 |
1 |
0.000626 |
| 10700 |
1 |
0.000625 |
| 10800 |
1 |
0.000625 |
| 10900 |
1 |
0.000626 |
| 11100 |
1 |
0.000625 |
| 11500 |
1 |
0.000625 |
| 11600 |
1 |
0.000625 |
| 11700 |
1 |
0.000625 |
| 11800 |
1 |
0.000625 |
| 12200 |
1 |
0.000625 |
| 12600 |
1 |
0.000625 |
| 13400 |
1 |
0.000625 |
| 14000 |
1 |
0.000625 |
| 14100 |
1 |
0.000625 |
| 14200 |
1 |
0.000626 |
| 14700 |
1 |
0.000625 |
| 15500 |
1 |
0.000625 |
| 16500 |
1 |
0.000625 |
| 16600 |
1 |
0.000625 |
| 16600 |
1 |
0.000625 |
| 17300 |
1 |
0.000625 |
| 17600 |
1 |
0.000625 |
| 17600 |
1 |
0.000625 |
| 17700 |
1 |
0.000625 |
| 18000 |
1 |
0.000625 |
| 18300 |
1 |
0.000625 |
| 18500 |
1 |
0.000625 |
| 18600 |
1 |
0.000625 |
| 18700 |
1 |
0.000625 |
| 19200 |
1 |
0.000625 |
| 19700 |
1 |
0.000626 |
| 19800 |
1 |
0.000626 |
| 26000 |
1 |
0.000626 |
| 26000 |
1 |
0.000626 |
| 26600 |
1 |
0.000626 |
| 27000 |
1 |
0.000626 |
| 29300 |
1 |
0.000626 |
| 29300 |
1 |
0.000626 |
| 29800 |
1 |
0.000626 |
| 30500 |
1 |
0.000626 |
| 31100 |
1 |
0.000626 |
| 34100 |
1 |
0.000626 |
| 35800 |
1 |
0.000626 |
| 36400 |
1 |
0.000626 |
| 36600 |
1 |
0.000626 |
| 36900 |
1 |
0.000626 |
| 38300 |
1 |
0.000626 |
| 38500 |
1 |
0.000626 |
| 38600 |
1 |
0.000626 |
| 38700 |
1 |
0.000626 |
| 42300 |
1 |
0.000626 |
| 42500 |
1 |
0.000626 |
| 43200 |
1 |
0.000626 |
| 45000 |
1 |
0.000626 |
| 45500 |
1 |
0.000626 |
| 45600 |
1 |
0.000626 |
| 46700 |
1 |
0.000626 |
| 48700 |
1 |
0.000626 |
| 49400 |
1 |
0.000626 |
| 49700 |
1 |
0.000626 |
| 50600 |
1 |
0.000626 |
| 51100 |
1 |
0.000626 |
| 51800 |
1 |
0.000626 |
| 52500 |
1 |
0.000626 |
| 52500 |
1 |
0.000626 |
| 53000 |
1 |
0.000626 |
| 53000 |
1 |
0.000626 |
| 59400 |
1 |
0.000626 |
| 60900 |
1 |
0.000626 |
| 62300 |
1 |
0.000626 |
| 67000 |
1 |
0.000626 |
| 67200 |
1 |
0.000626 |
| 67300 |
1 |
0.000626 |
| 68400 |
1 |
0.000626 |
| 68500 |
1 |
0.000626 |
| 68700 |
1 |
0.000626 |
| 69000 |
1 |
0.000626 |
| 69100 |
1 |
0.000626 |
| 69400 |
1 |
0.000626 |
| 69800 |
1 |
0.000626 |
| 70700 |
1 |
0.000626 |
| 70800 |
1 |
0.000626 |
| 71000 |
1 |
0.000626 |
|
CL4 ----> CL40
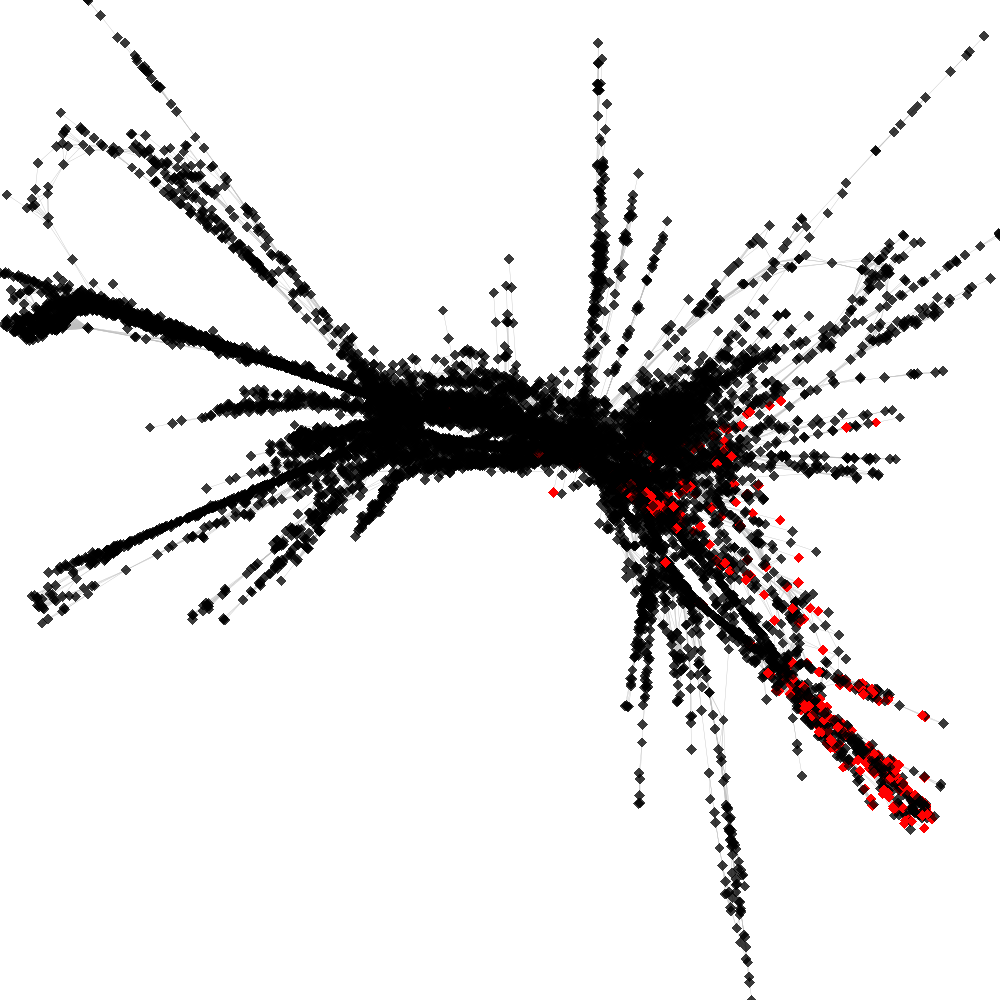
No. of shared pairs: :343
CL4 ----> CL32
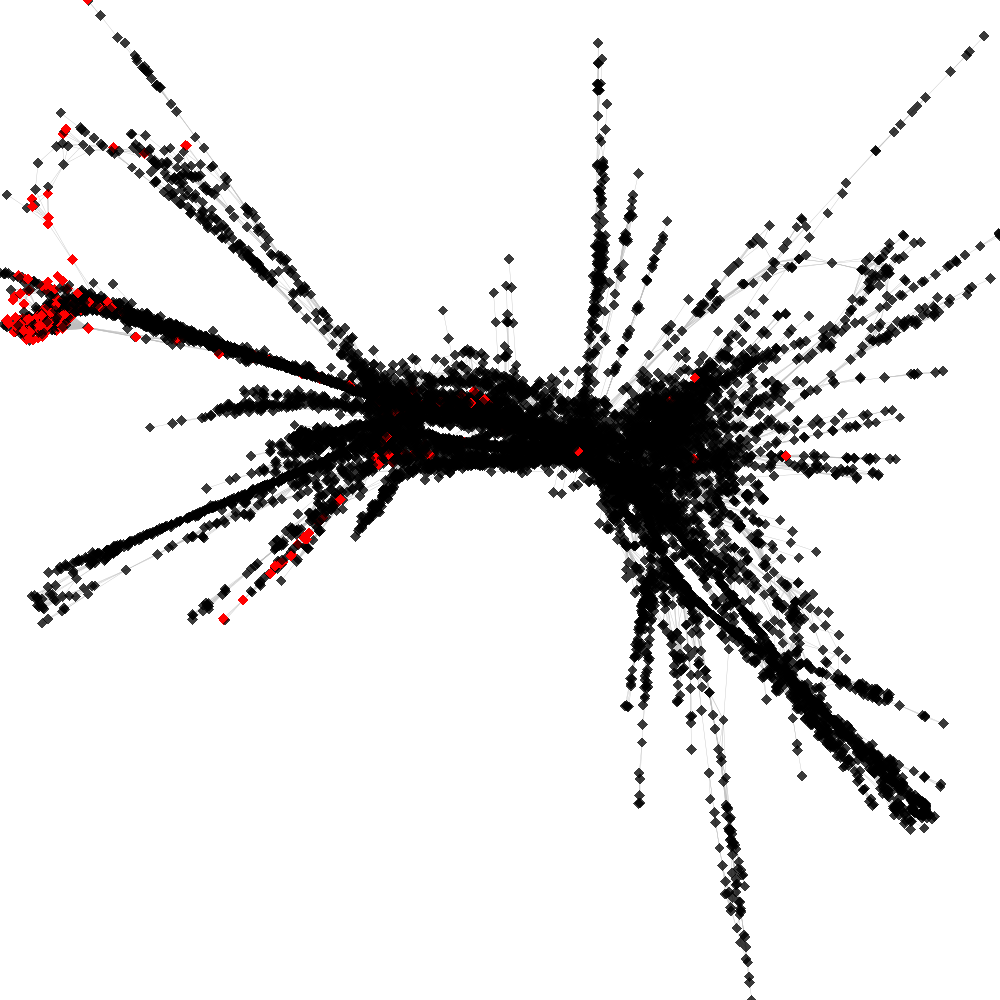
No. of shared pairs: :257
CL4 ----> CL167
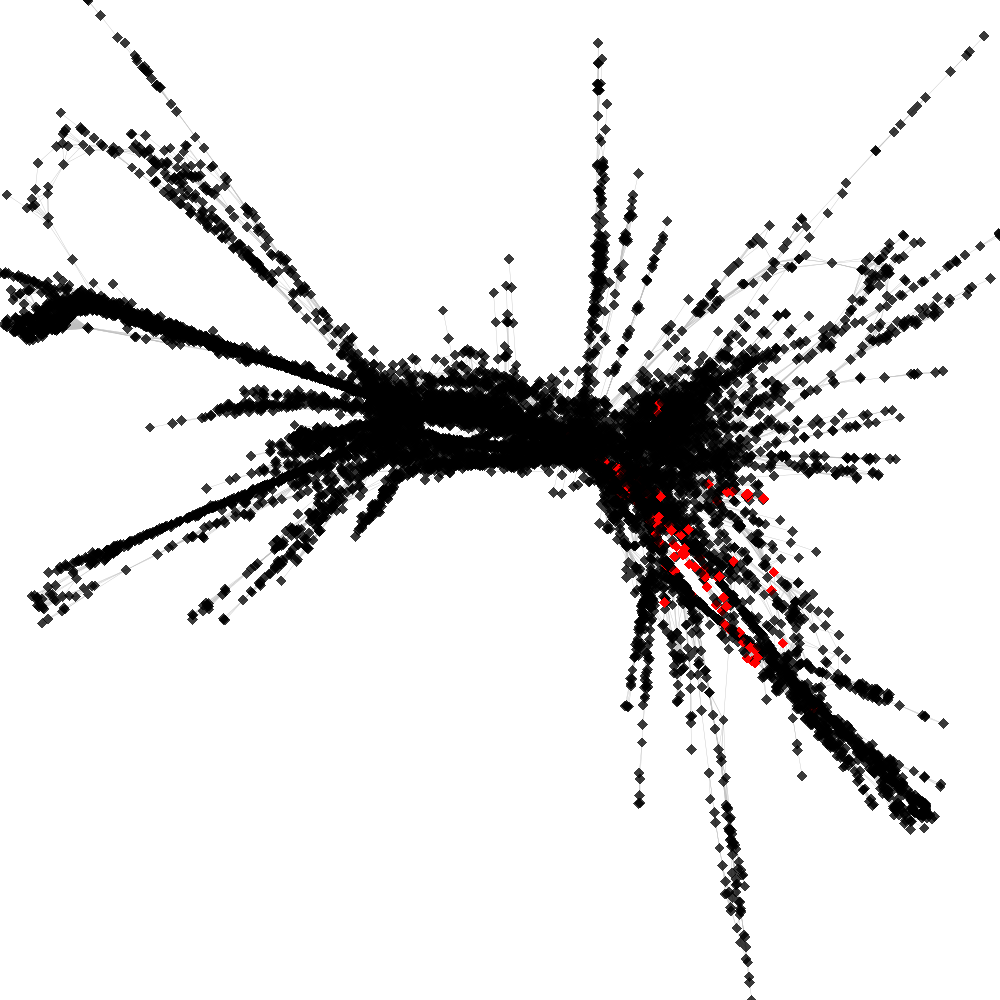
No. of shared pairs: :144
CL4 ----> CL114
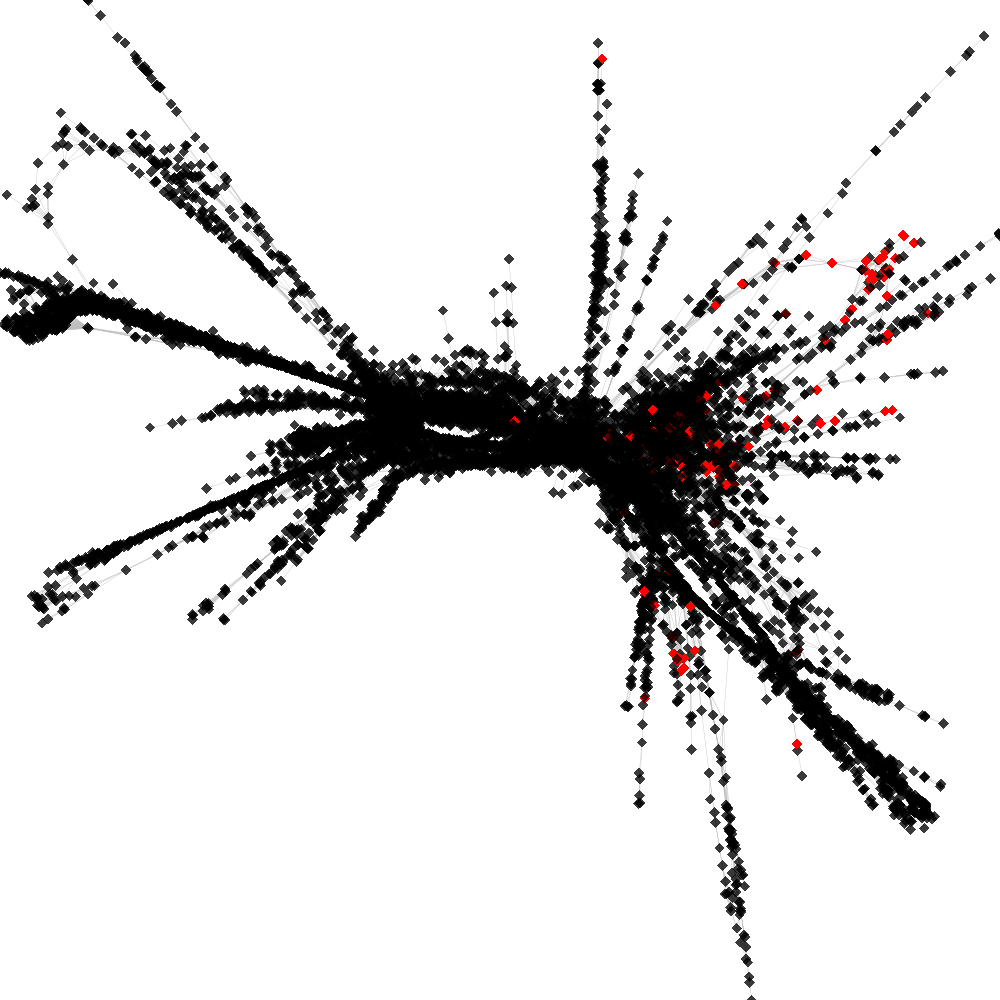
No. of shared pairs: :140
CL4 ----> CL79
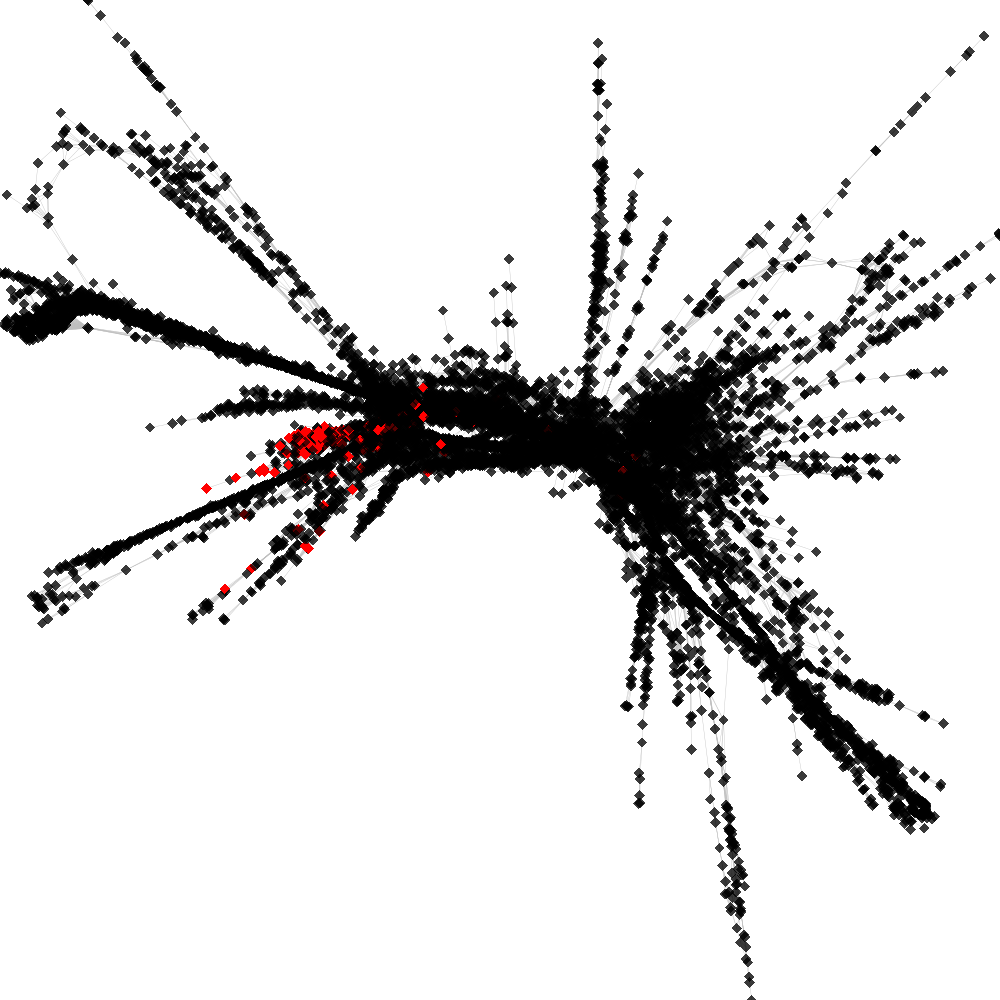
No. of shared pairs: :134
CL4 ----> CL104
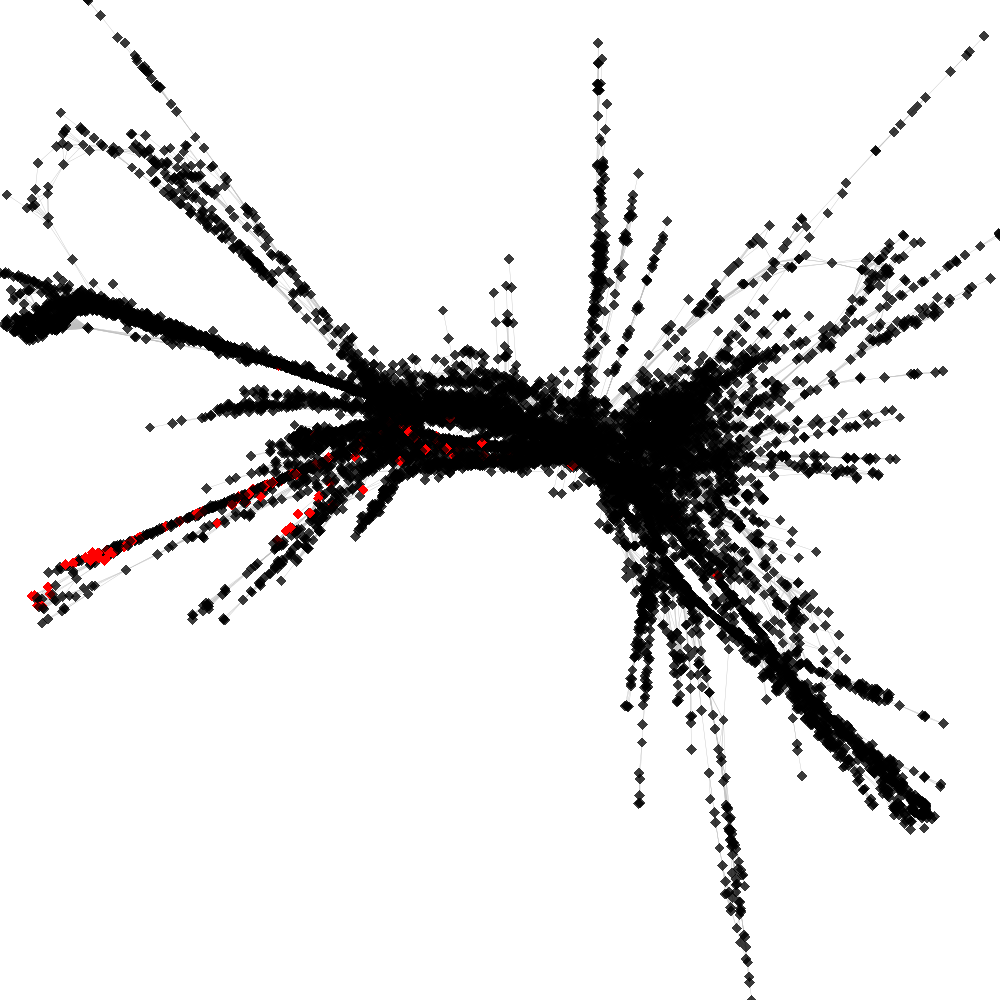
No. of shared pairs: :116
CL4 ----> CL166
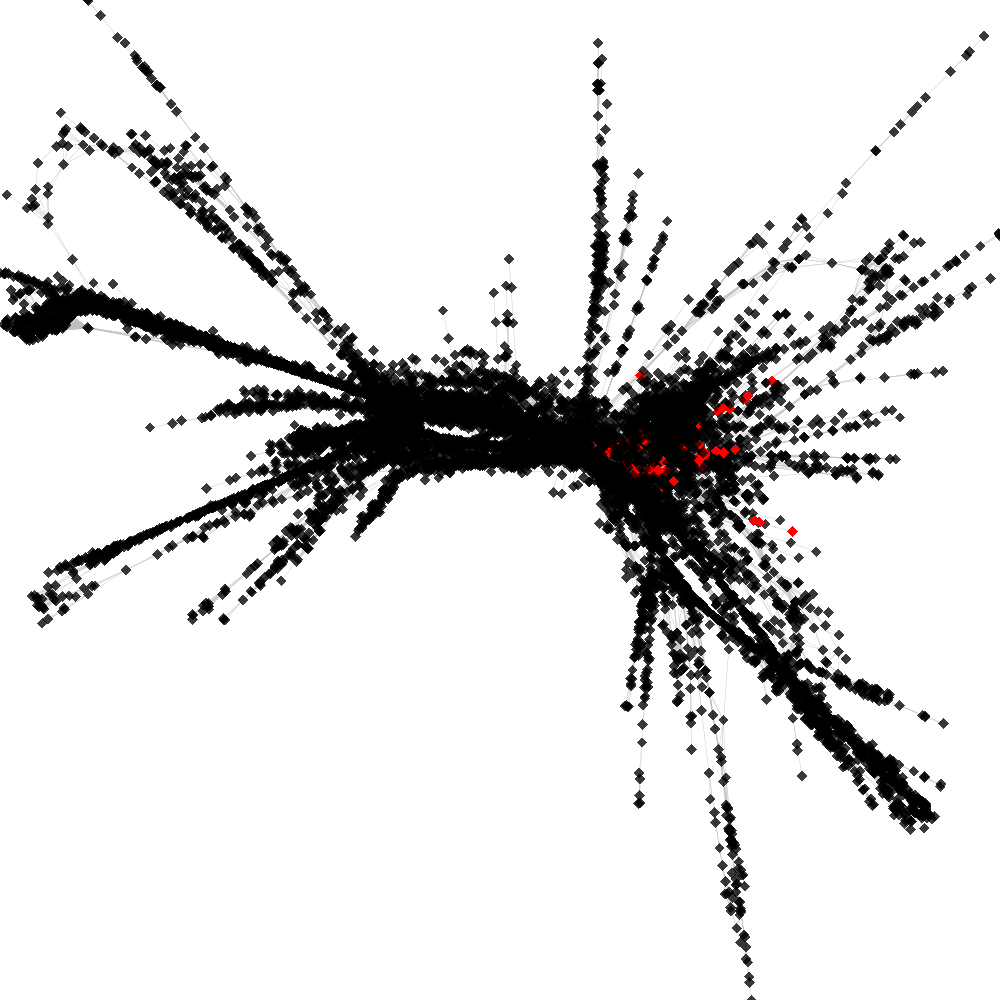
No. of shared pairs: :108
CL4 ----> CL103
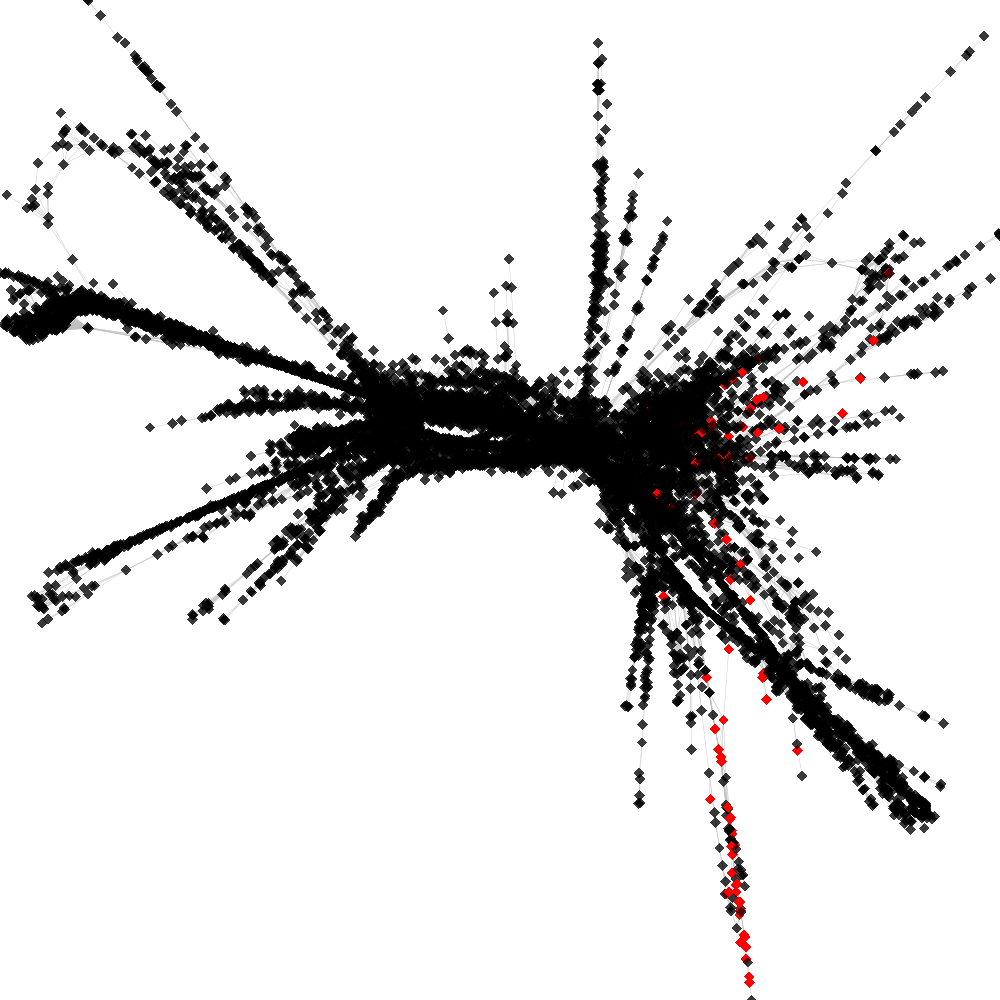
No. of shared pairs: :92
CL4 ----> CL203
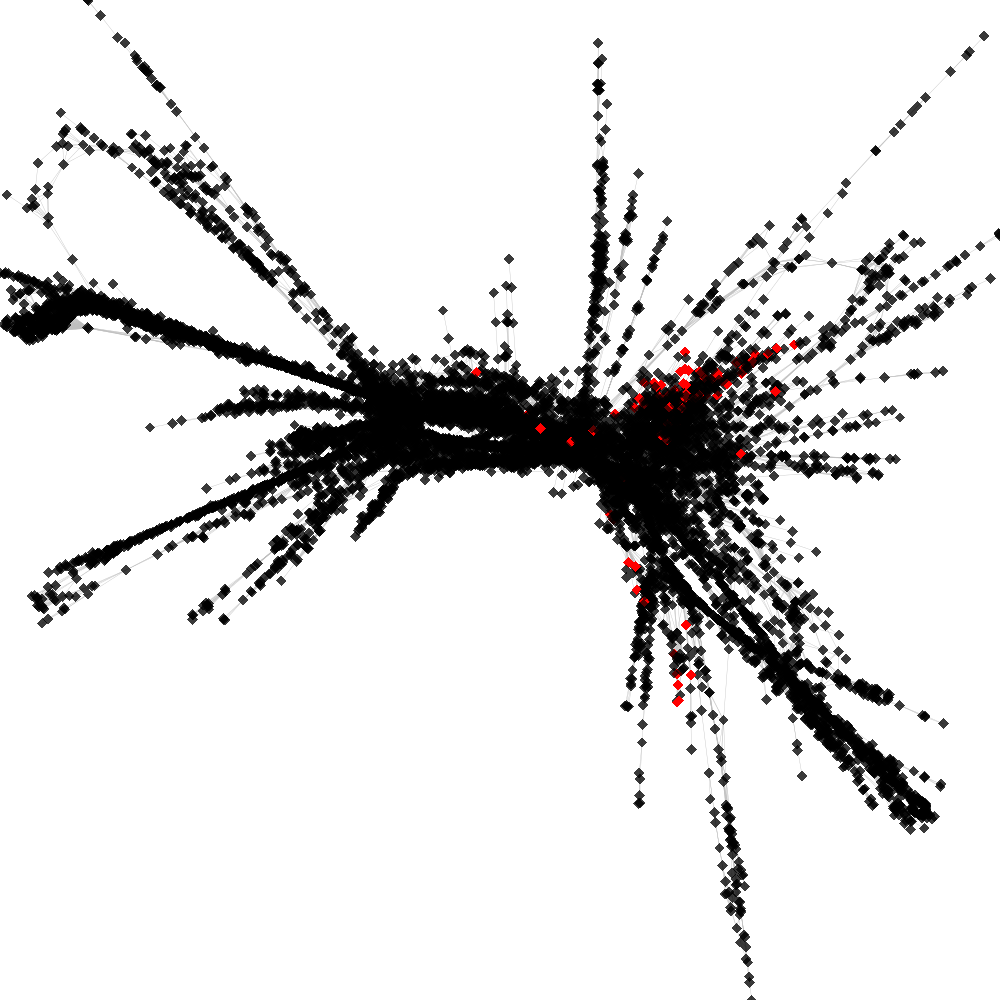
No. of shared pairs: :86
CL4 ----> CL9
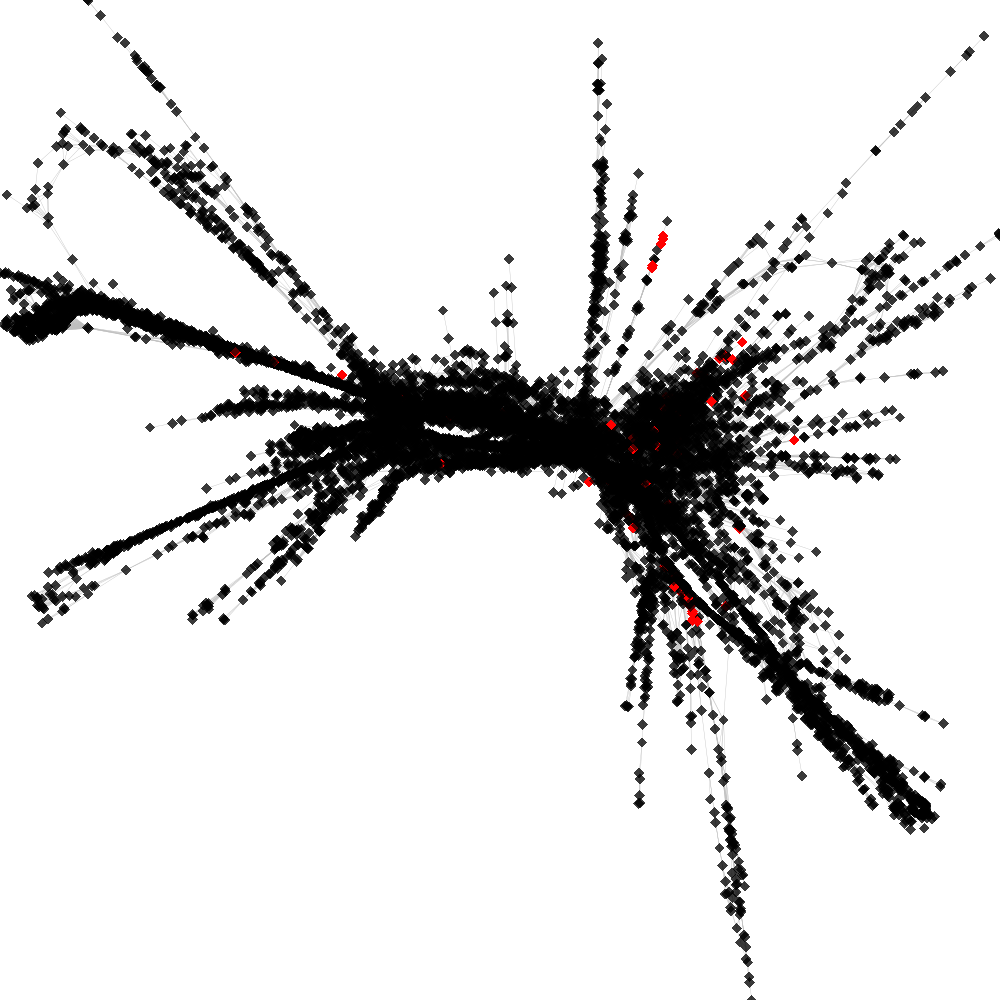
No. of shared pairs: :82
CL4 ----> CL53
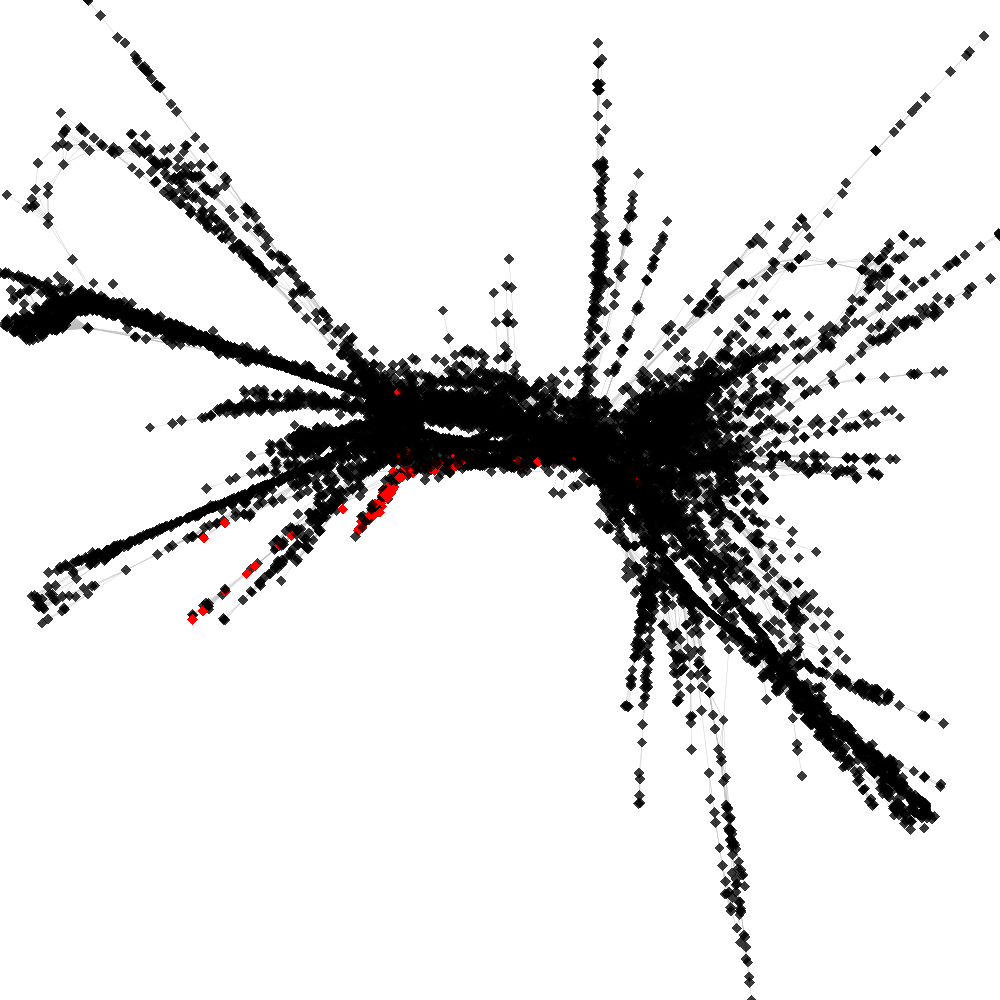
No. of shared pairs: :78
CL4 ----> CL221

No. of shared pairs: :52
CL4 ----> CL209

No. of shared pairs: :43
CL4 ----> CL12
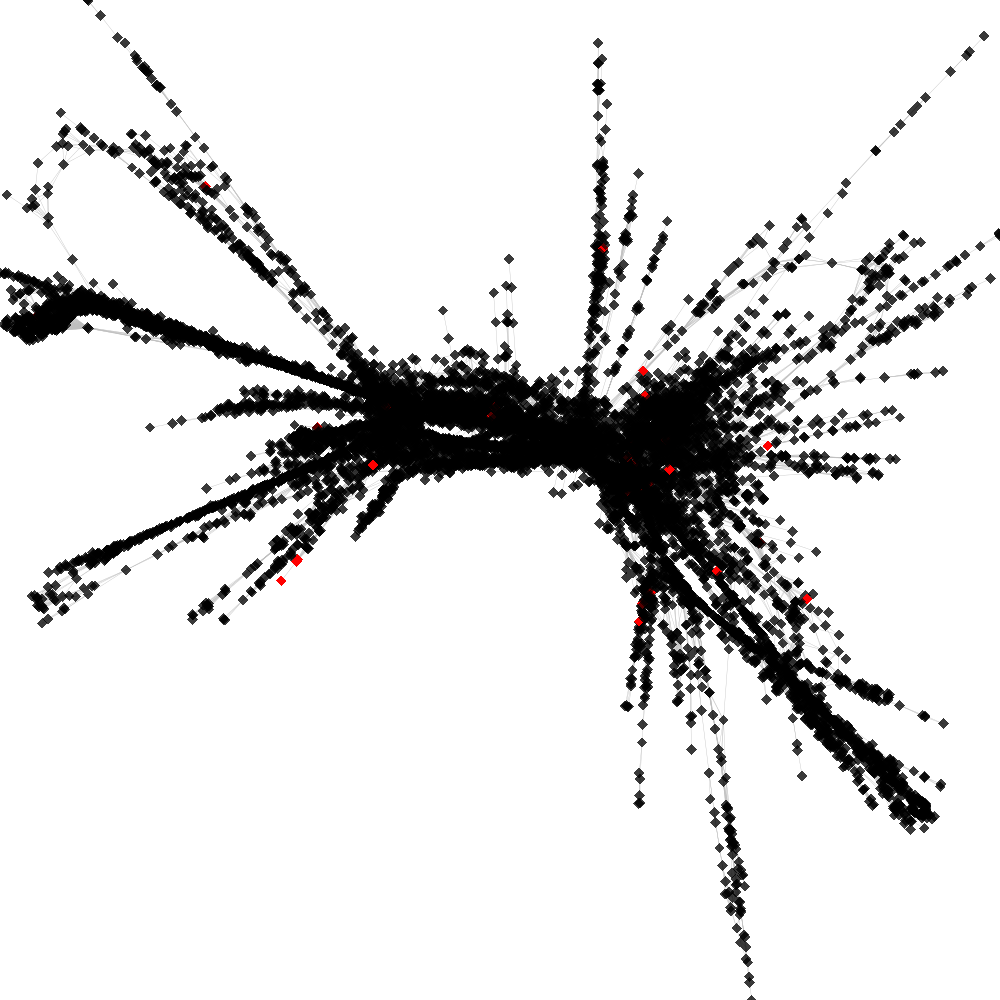
No. of shared pairs: :36
CL4 ----> CL213
No. of shared pairs: :36
CL4 ----> CL75
No. of shared pairs: :25
CL4 ----> CL11
No. of shared pairs: :22
CL4 ----> CL287

No. of shared pairs: :19
CL4 ----> CL5

No. of shared pairs: :17
CL4 ----> CL247

No. of shared pairs: :16
CL4 ----> CL8

No. of shared pairs: :15
CL4 ----> CL22

No. of shared pairs: :12


