| size | 920 |
| size_real | 920 |
| ecount | 4553 |
| supercluster | 34 |
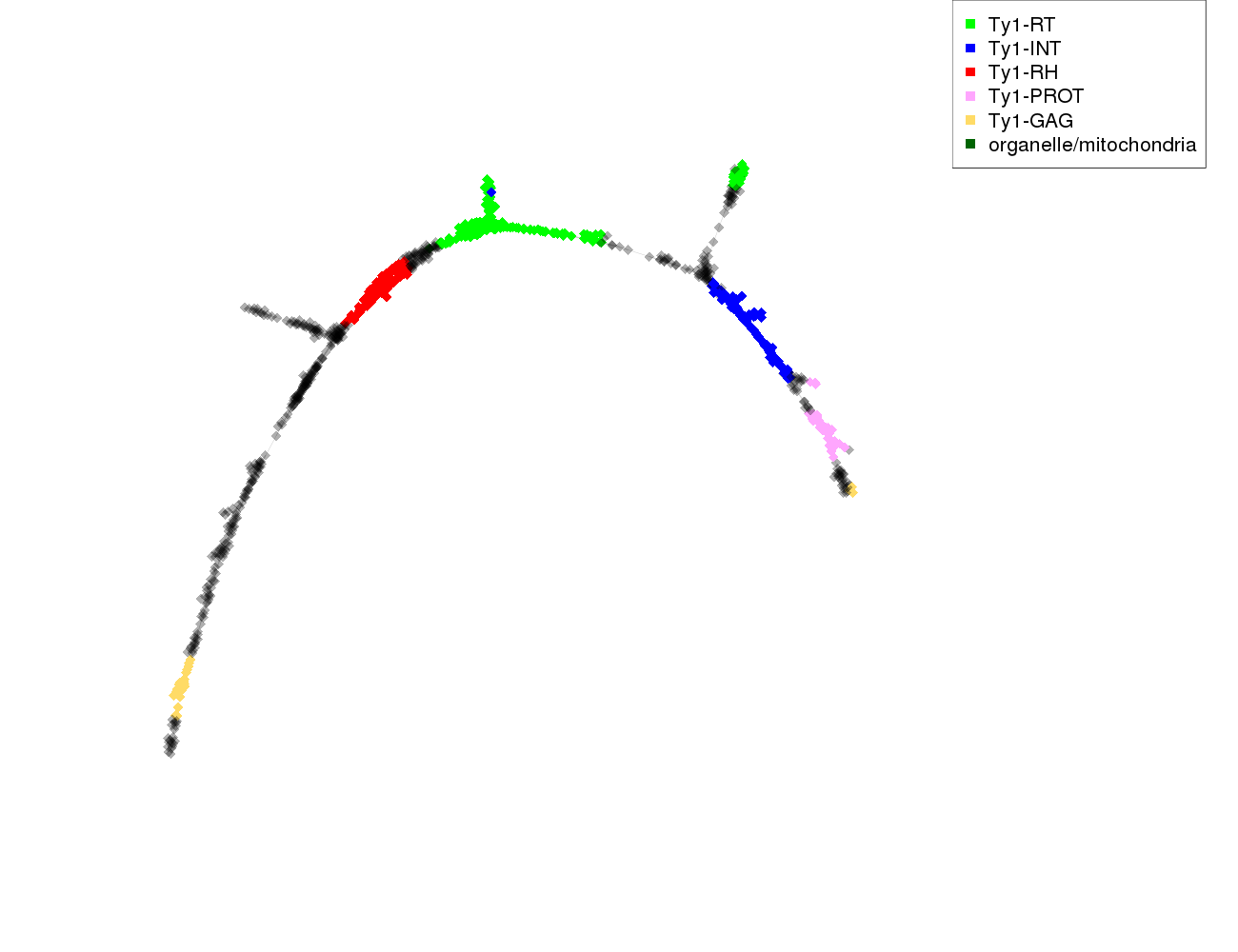
| annotations_summary | 19.13% Class_I/LTR/Ty1_copia/TAR:Ty1-RT 12.17% Class_I/LTR/Ty1_copia/TAR:Ty1-INT 11.41% Class_I/LTR/Ty1_copia/TAR:Ty1-RH 3.48% Class_I/LTR/Ty1_copia/TAR:Ty1-PROT 2.83% Class_I/LTR/Ty1_copia/TAR:Ty1-GAG 0.11% organelle/mitochondria 0.11% Class_I/LTR/Ty1_copia/Ikeros:Ty1-RT |
| pair_completeness | 0.762452107279693 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.00108695652173913 |
| satellite_probability | 5.68788576307886e-23 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S8 | 316 |
| S9 | 300 |
| S7 | 304 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 613 | 556 | 510 |
| S9 | 556 | 464 | 458 |
| S7 | 510 | 458 | 428 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 0.991 | 1.020 | 0.991 |
| S9 | 1.020 | 0.967 | 1.010 |
| S7 | 0.991 | 1.010 | 0.999 |
protein domains:
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|