| size | 2086 |
| size_real | 2086 |
| ecount | 12488 |
| supercluster | 17 |
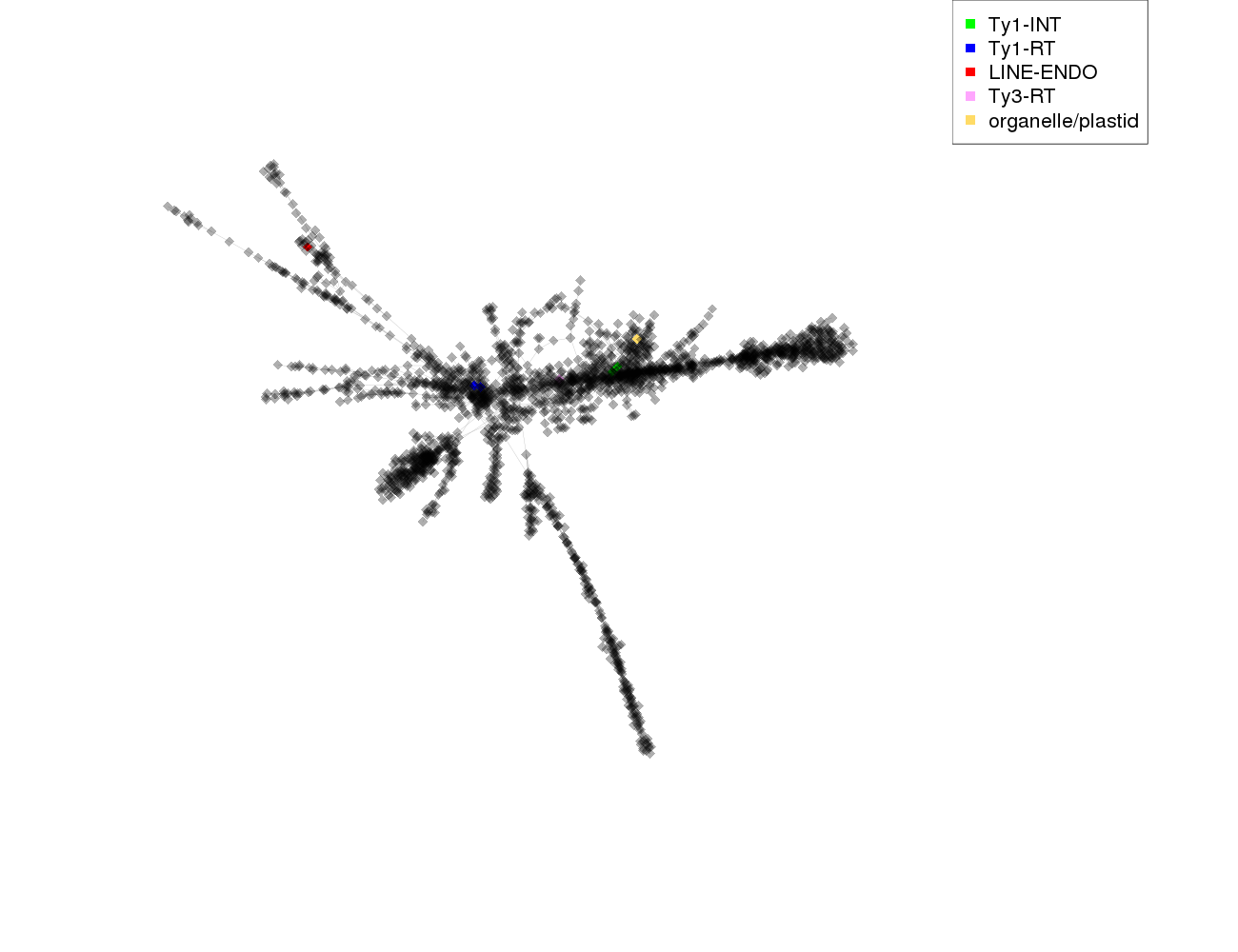
| annotations_summary | 0.10% Class_I/LTR/Ty1_copia/Tork:Ty1-RT 0.10% Class_I/LTR/Ty1_copia/Ale:Ty1-INT 0.05% Class_I/LINE:LINE-ENDO 0.05% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-RT 0.05% organelle/plastid |
| pair_completeness | 0.353666450356911 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.000479386385426654 |
| satellite_probability | 9.19560910382493e-21 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
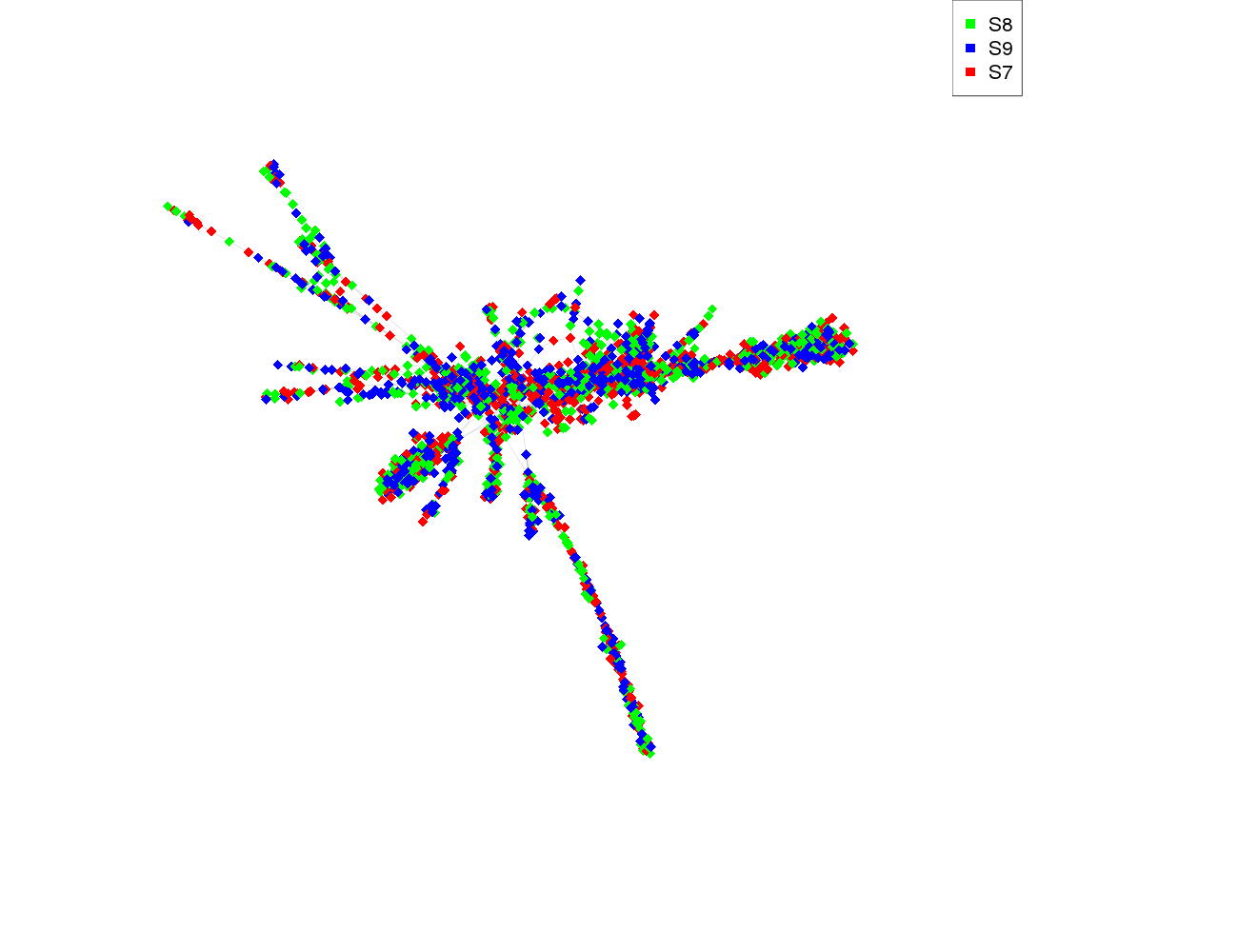
| S8 | 687 |
| S9 | 689 |
| S7 | 710 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 1360 | 1340 | 1400 |
| S9 | 1340 | 1380 | 1400 |
| S7 | 1400 | 1400 | 1450 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 1.010 | 0.989 | 1.000 |
| S9 | 0.989 | 1.020 | 0.996 |
| S7 | 1.000 | 0.996 | 1.000 |
protein domains:
|
|
||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|