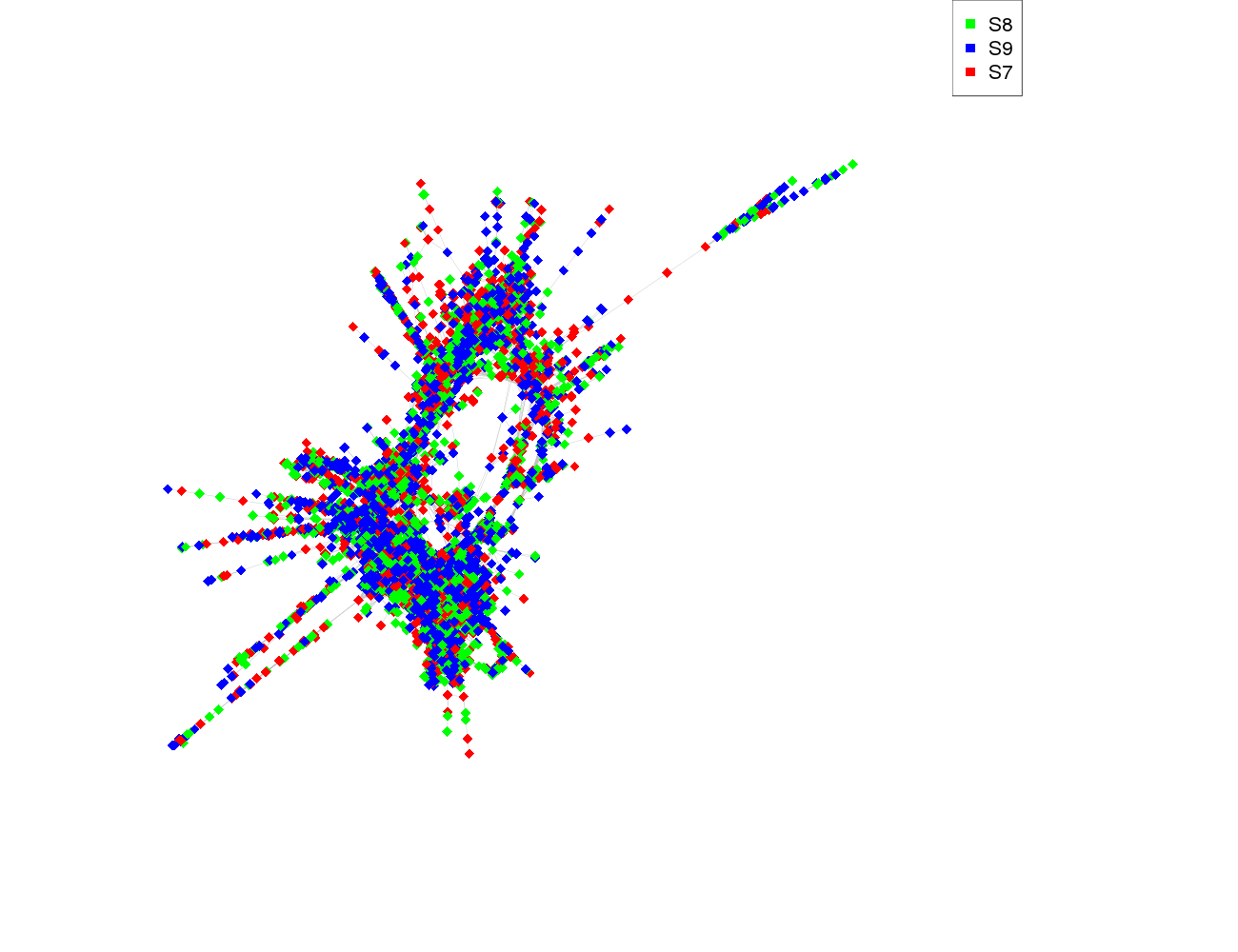
| size | 5713 |
| size_real | 5713 |
| ecount | 93641 |
| supercluster | 3 |
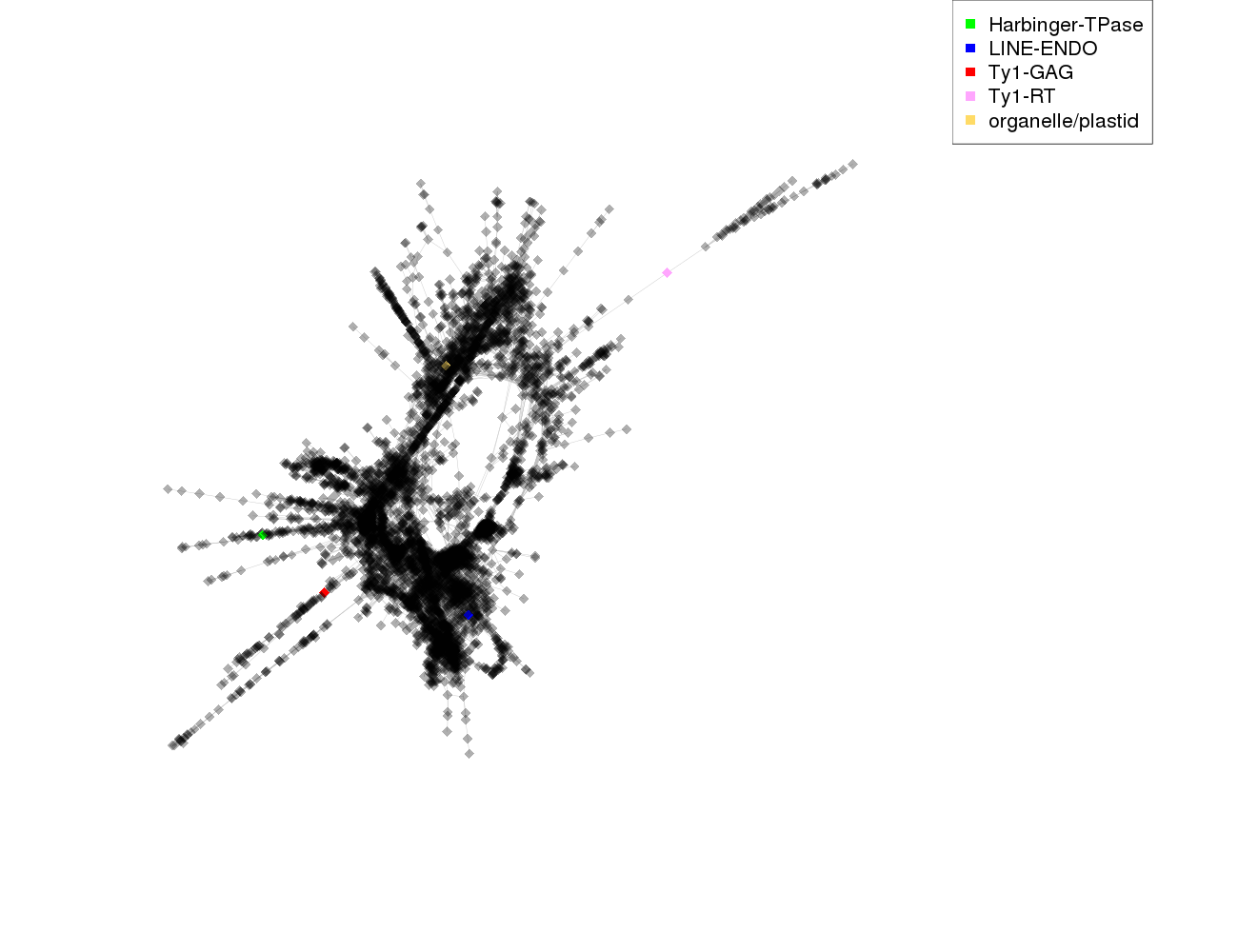
| annotations_summary | 0.02% Class_I/LINE:LINE-ENDO 0.02% Class_I/LTR/Ty1_copia/Ale:Ty1-RT 0.02% organelle/plastid 0.02% Class_I/LTR/Ty1_copia/Alesia:Ty1-GAG 0.02% Class_II/Subclass_1/TIR/PIF_Harbinger:Harbinger-TPase |
| pair_completeness | 0.50818373812038 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.404691055487485 |
| satellite_probability | 6.11456497571856e-07 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S8 | 1910 |
| S9 | 1920 |
| S7 | 1880 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 11600 | 10800 | 10400 |
| S9 | 10800 | 10300 | 9750 |
| S7 | 10400 | 9750 | 9850 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 1.010 | 0.998 | 0.994 |
| S9 | 0.998 | 1.020 | 0.986 |
| S7 | 0.994 | 0.986 | 1.020 |
protein domains:
|
|
||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|