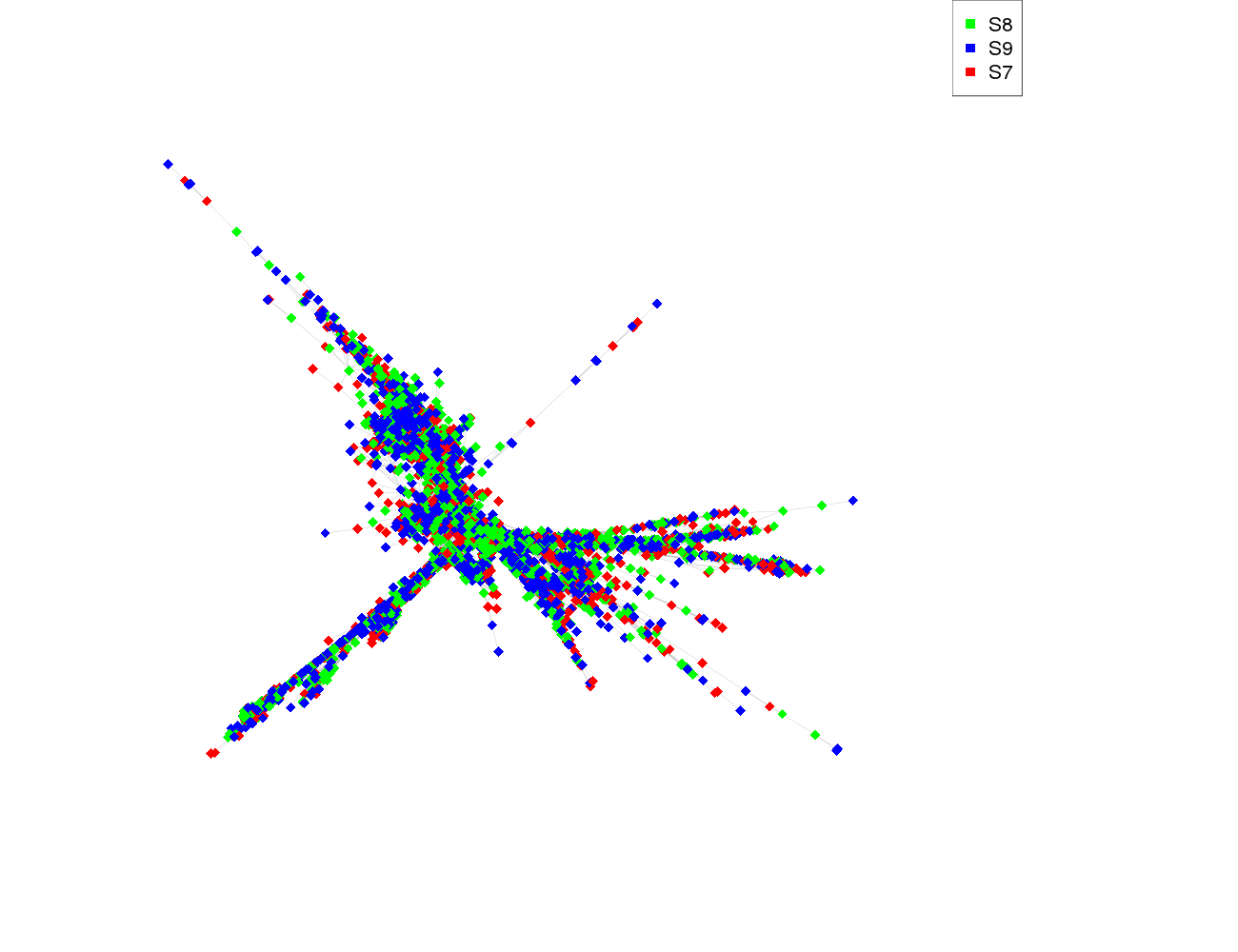
| size | 6275 |
| size_real | 6275 |
| ecount | 466169 |
| supercluster | 1 |
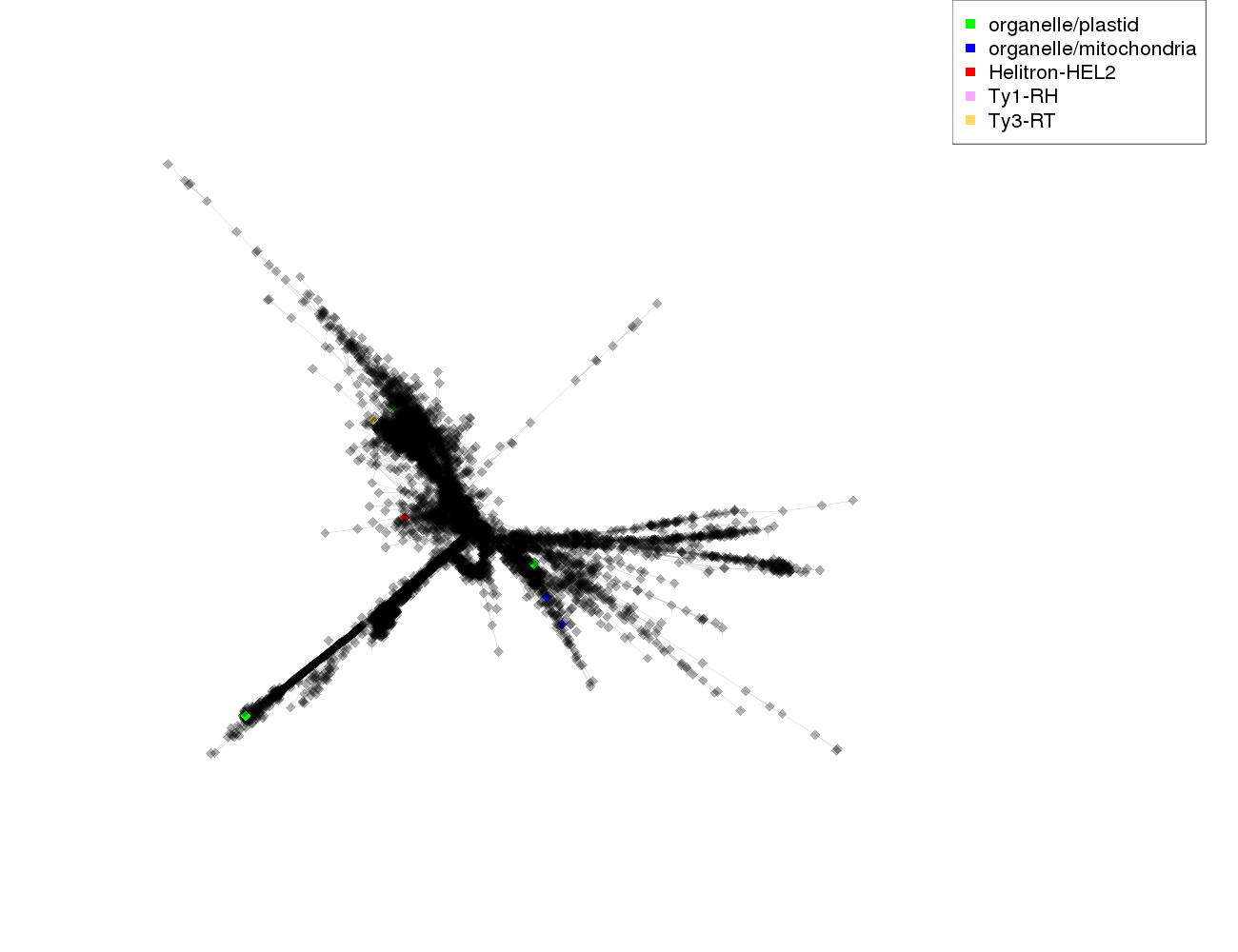
| annotations_summary | 0.05% organelle/plastid 0.03% organelle/mitochondria 0.02% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-RT 0.02% Class_II/Subclass_2/Helitron:Helitron-HEL2 0.02% Class_I/LTR/Ty1_copia/Tork:Ty1-RH |
| pair_completeness | 0.366804617730342 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.00828685258964143 |
| satellite_probability | 1.14691457676211e-20 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S8 | 2070 |
| S9 | 2100 |
| S7 | 2100 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 52400 | 51600 | 52500 |
| S9 | 51600 | 50800 | 51500 |
| S7 | 52500 | 51500 | 51900 |
| S8 | S9 | S7 | |
|---|---|---|---|
| S8 | 0.998 | 0.999 | 1.000 |
| S9 | 0.999 | 1.000 | 1.000 |
| S7 | 1.000 | 1.000 | 0.996 |
protein domains:
|
|
||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|