Cluster no. 1
Go back to cluster table
Cluster is part of supercluster: 2
Cluster characteristics:
| size |
17937 |
| size_real |
17937 |
| ecount |
904564 |
| supercluster |
2 |
| annotations_summary |
4.40% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-CHDII
0.12% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-INT
0.07% organelle/plastid
0.03% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-INT
0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIV_Ogre:Ty3-RH
0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-RH
0.01% organelle/mitochondria
0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-PROT
0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-RT
0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIII:Ty3-RH
0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RT
0.01% Class_I/LTR/Ty1_copia/Ivana:Ty1-RH
0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Tcn1:Ty3-RT
0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-PROT
0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Galadriel:Ty3-RH
0.01% Class_I/LTR/Ty1_copia/SIRE:Ty1-INT
0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-GAG
0.01% Class_I/LTR/Ty1_copia/Ale:Ty1-RT
0.01% Class_I/LTR/Ty1_copia/Ale:Ty1-RH
|
| pair_completeness |
0.537413216765235 |
| pbs_score |
None |
| TR_score |
None |
| TR_monomer_length |
None |
| loop_index |
0.00111501365891732 |
| satellite_probability |
1.65508917325132e-21 |
| consensus |
None |
| TAREAN_annotation |
Other |
| orientation_score |
1 |
comparative analysis:
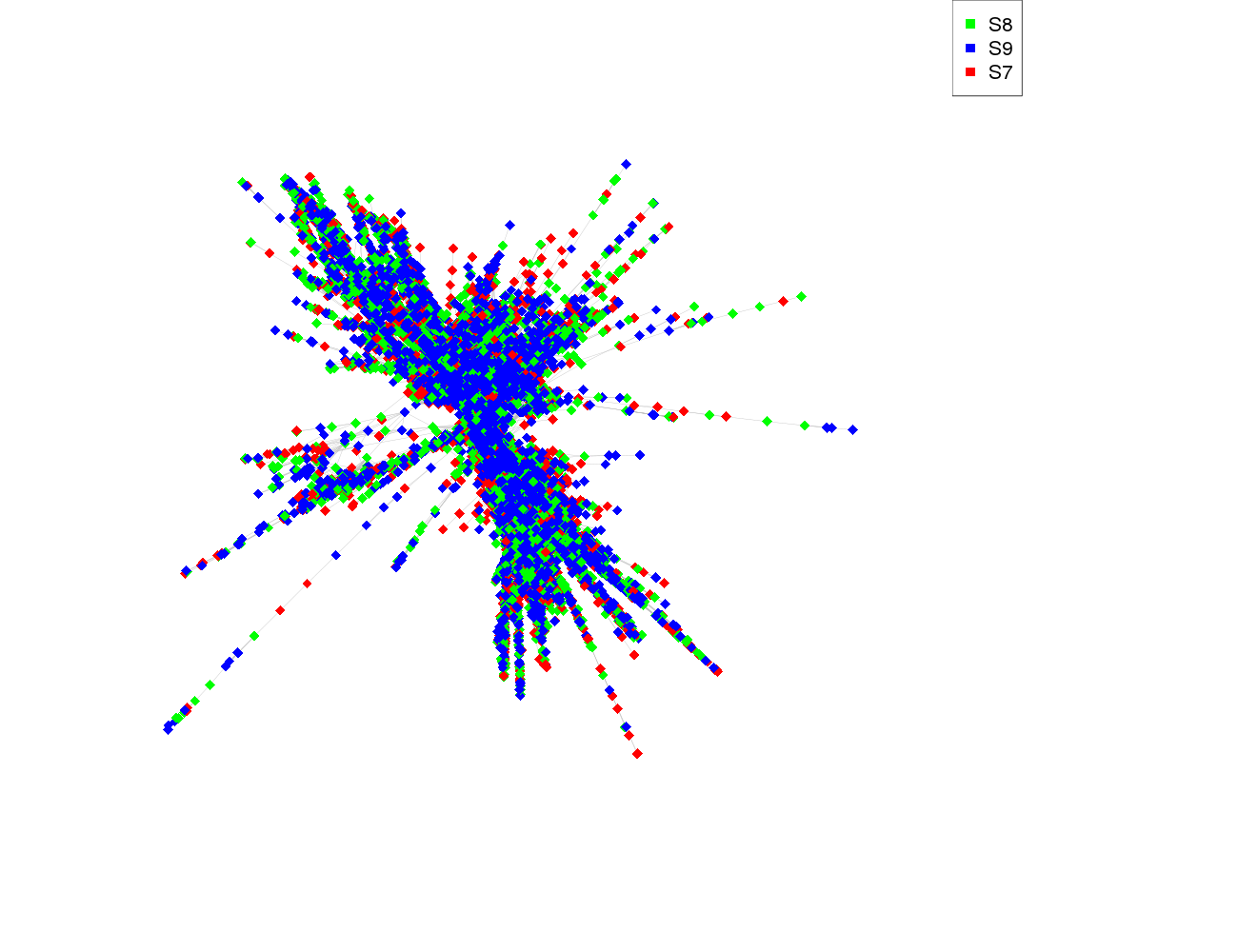
Comparative analysis - species read counts:
| Species |
Read count |
| S8 |
5950 |
| S9 |
6160 |
| S7 |
5830 |
comparative analysis - number of edges between species:
|
S8 |
S9 |
S7 |
| S8 |
97600 |
105000 |
94700 |
| S9 |
105000 |
113000 |
102000 |
| S7 |
94700 |
102000 |
92300 |
comparative analysis - observed/expected number of edges between species
|
S8 |
S9 |
S7 |
| S8 |
1.000 |
0.999 |
1.000 |
| S9 |
0.999 |
1.000 |
0.999 |
| S7 |
1.000 |
0.999 |
1.000 |
protein domains:
protein domains:
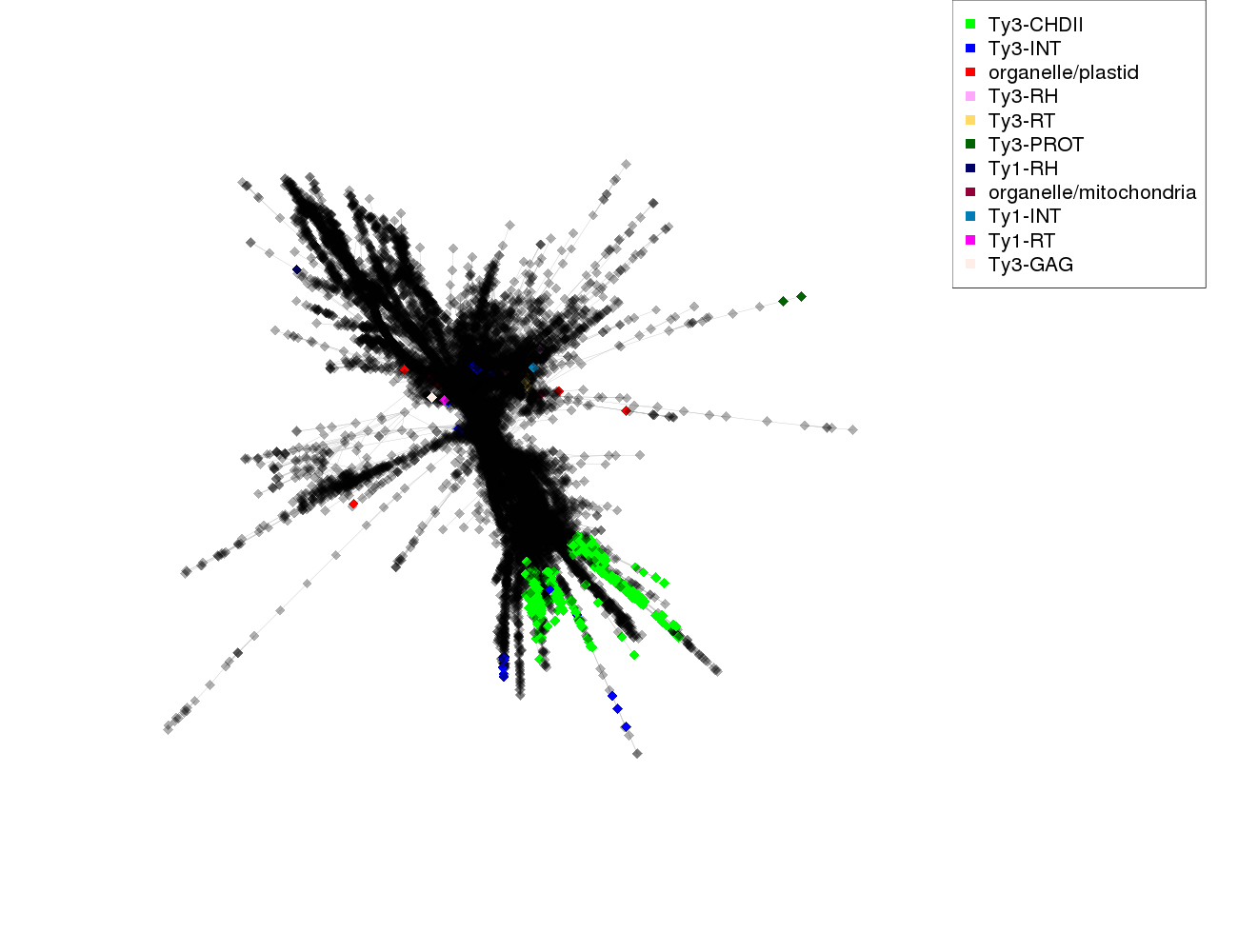
Reads annotation summary
| |
cl_string |
domain |
Freq |
proportion |
| mitochondria
|
mitochondria
|
|
2
|
1.1e-04
|
| plastid
|
plastid
|
|
13
|
7.2e-04
|
| SIRE Ty1-INT
|
SIRE
|
Ty1-INT
|
1
|
5.6e-05
|
| Ale Ty1-RH
|
Ale
|
Ty1-RH
|
1
|
5.6e-05
|
| Ivana Ty1-RH
|
Ivana
|
Ty1-RH
|
1
|
5.6e-05
|
| Ale Ty1-RT
|
Ale
|
Ty1-RT
|
1
|
5.6e-05
|
| Tekay Ty3-CHDII
|
Tekay
|
Ty3-CHDII
|
790
|
4.4e-02
|
| TatV Ty3-GAG
|
TatV
|
Ty3-GAG
|
1
|
5.6e-05
|
| Reina Ty3-INT
|
Reina
|
Ty3-INT
|
5
|
2.8e-04
|
| Tekay Ty3-INT
|
Tekay
|
Ty3-INT
|
22
|
1.2e-03
|
| Athila Ty3-PROT
|
Athila
|
Ty3-PROT
|
1
|
5.6e-05
|
| TatV Ty3-PROT
|
TatV
|
Ty3-PROT
|
2
|
1.1e-04
|
| Galadriel Ty3-RH
|
Galadriel
|
Ty3-RH
|
1
|
5.6e-05
|
| TatIII Ty3-RH
|
TatIII
|
Ty3-RH
|
1
|
5.6e-05
|
| TatIV_Ogre Ty3-RH
|
TatIV_Ogre
|
Ty3-RH
|
3
|
1.7e-04
|
| TatV Ty3-RH
|
TatV
|
Ty3-RH
|
3
|
1.7e-04
|
| TatV Ty3-RT
|
TatV
|
Ty3-RT
|
2
|
1.1e-04
|
| Tcn1 Ty3-RT
|
Tcn1
|
Ty3-RT
|
1
|
5.6e-05
|
| Tekay Ty3-RT
|
Tekay
|
Ty3-RT
|
1
|
5.6e-05
|
|
clusters with similarity:
| Cluster |
Number of similarity hits |
|
| 46 |
3790 |
| 14 |
1560 |
| 45 |
1370 |
| 7 |
1150 |
| 62 |
492 |
| 34 |
450 |
| 87 |
304 |
| 51 |
293 |
| 65 |
162 |
| 10 |
138 |
| 28 |
132 |
| 54 |
125 |
| 89 |
104 |
| 22 |
73 |
| 90 |
52 |
| 66 |
51 |
| 75 |
43 |
| 105 |
40 |
| 42 |
36 |
| 67 |
22 |
| 37 |
17 |
| 102 |
14 |
| 71 |
10 |
| 200 |
8 |
| 218 |
3 |
| 58 |
2 |
| 6 |
1 |
| 9 |
1 |
| 12 |
1 |
| 44 |
1 |
| 127 |
1 |
| 343 |
1 |
| 388 |
1 |
|
clusters connected through mates:
| Cluster |
Number of shared
read pairs |
k |
|
| 14 |
377 |
0.106 |
| 7 |
349 |
0.0983 |
| 46 |
330 |
0.107 |
| 45 |
212 |
0.0714 |
| 62 |
181 |
0.0608 |
| 28 |
178 |
0.0554 |
| 51 |
154 |
0.0519 |
| 34 |
151 |
0.0507 |
| 65 |
149 |
0.0504 |
| 75 |
147 |
0.0497 |
| 54 |
139 |
0.0464 |
| 22 |
118 |
0.0287 |
| 66 |
111 |
0.0374 |
| 87 |
111 |
0.0391 |
| 10 |
100 |
0.0282 |
| 67 |
85 |
0.0294 |
| 71 |
83 |
0.0281 |
| 37 |
70 |
0.0212 |
| 15 |
58 |
0.0154 |
| 42 |
49 |
0.0162 |
| 105 |
37 |
0.0134 |
| 89 |
36 |
0.013 |
| 2 |
28 |
0.006 |
| 12 |
28 |
0.00775 |
| 90 |
28 |
0.00985 |
| 200 |
24 |
0.00882 |
| 3 |
23 |
0.00403 |
| 127 |
23 |
0.00838 |
| 5 |
21 |
0.00392 |
| 13 |
20 |
0.00403 |
| 17 |
18 |
0.00504 |
| 23 |
17 |
0.00467 |
| 102 |
17 |
0.00613 |
| 6 |
15 |
0.00287 |
| 16 |
15 |
0.00319 |
| 8 |
13 |
0.00312 |
| 21 |
12 |
0.00358 |
| 24 |
12 |
0.00309 |
| 25 |
12 |
0.00386 |
| 36 |
12 |
0.00359 |
| 218 |
12 |
0.00441 |
| 4 |
9 |
0.00174 |
| 19 |
9 |
0.00196 |
| 33 |
9 |
0.0029 |
| 55 |
9 |
0.00304 |
| 18 |
8 |
0.00218 |
| 30 |
8 |
0.00241 |
| 38 |
8 |
0.00225 |
| 39 |
8 |
0.00228 |
| 41 |
8 |
0.00218 |
| 44 |
8 |
0.00265 |
| 58 |
8 |
0.00286 |
| 26 |
7 |
0.00158 |
| 53 |
7 |
0.00233 |
| 80 |
7 |
0.00239 |
| 11 |
6 |
0.00131 |
| 20 |
6 |
0.00131 |
| 68 |
6 |
0.00205 |
| 73 |
6 |
0.00201 |
| 27 |
5 |
0.0012 |
| 64 |
5 |
0.00165 |
| 69 |
5 |
0.00173 |
| 388 |
5 |
0.00185 |
| 9 |
4 |
0.000836 |
| 29 |
4 |
0.00101 |
| 31 |
4 |
0.00118 |
| 35 |
4 |
0.0011 |
| 40 |
4 |
0.00123 |
| 63 |
4 |
0.00123 |
| 76 |
4 |
0.00137 |
| 320 |
4 |
0.00148 |
| 830 |
4 |
0.00148 |
| 977 |
4 |
0.00148 |
| 1280 |
4 |
0.00148 |
| 47 |
3 |
0.00104 |
| 48 |
3 |
0.000845 |
| 79 |
3 |
0.00104 |
| 84 |
3 |
0.00102 |
| 1260 |
3 |
0.00111 |
| 1570 |
3 |
0.00111 |
| 2640 |
3 |
0.00111 |
| 5970 |
3 |
0.00111 |
| 32 |
2 |
0.000475 |
| 43 |
2 |
0.000593 |
| 50 |
2 |
0.000684 |
| 52 |
2 |
0.000534 |
| 60 |
2 |
0.000663 |
| 70 |
2 |
0.000667 |
| 77 |
2 |
0.00072 |
| 78 |
2 |
0.000689 |
| 95 |
2 |
0.000721 |
| 111 |
2 |
0.000726 |
| 115 |
2 |
0.00073 |
| 177 |
2 |
0.000736 |
| 179 |
2 |
0.000736 |
| 301 |
2 |
0.000737 |
| 481 |
2 |
0.00074 |
| 720 |
2 |
0.00074 |
| 1150 |
2 |
0.00074 |
| 1330 |
2 |
0.000741 |
| 1430 |
2 |
0.00074 |
| 1730 |
2 |
0.000741 |
| 1740 |
2 |
0.00074 |
| 2180 |
2 |
0.00074 |
| 2370 |
2 |
0.000741 |
| 2500 |
2 |
0.00074 |
| 4600 |
2 |
0.000741 |
| 5570 |
2 |
0.000741 |
| 6140 |
2 |
0.000741 |
| 9310 |
2 |
0.000741 |
| 9800 |
2 |
0.000741 |
| 10000 |
2 |
0.000741 |
| 14200 |
2 |
0.000741 |
| 16200 |
2 |
0.000741 |
| 17400 |
2 |
0.000741 |
| 19600 |
2 |
0.000741 |
| 19900 |
2 |
0.000741 |
| 22200 |
2 |
0.000741 |
| 25600 |
2 |
0.000741 |
| 56 |
1 |
0.000299 |
| 59 |
1 |
0.00029 |
| 61 |
1 |
0.000313 |
| 74 |
1 |
0.000329 |
| 81 |
1 |
0.000309 |
| 83 |
1 |
0.000332 |
| 85 |
1 |
0.000362 |
| 86 |
1 |
0.000345 |
| 88 |
1 |
0.000362 |
| 91 |
1 |
0.000354 |
| 92 |
1 |
0.000358 |
| 93 |
1 |
0.000346 |
| 99 |
1 |
0.000366 |
| 109 |
1 |
0.000368 |
| 112 |
1 |
0.000363 |
| 114 |
1 |
0.000362 |
| 120 |
1 |
0.00037 |
| 121 |
1 |
0.000367 |
| 122 |
1 |
0.000369 |
| 126 |
1 |
0.000365 |
| 128 |
1 |
0.000365 |
| 130 |
1 |
0.000366 |
| 134 |
1 |
0.000364 |
| 143 |
1 |
0.00037 |
| 150 |
1 |
0.000364 |
| 154 |
1 |
0.000365 |
| 158 |
1 |
0.000368 |
| 175 |
1 |
0.000367 |
| 187 |
1 |
0.000367 |
| 214 |
1 |
0.000369 |
| 222 |
1 |
0.000368 |
| 237 |
1 |
0.000369 |
| 255 |
1 |
0.000369 |
| 257 |
1 |
0.000368 |
| 262 |
1 |
0.000369 |
| 263 |
1 |
0.000369 |
| 286 |
1 |
0.000369 |
| 287 |
1 |
0.000369 |
| 302 |
1 |
0.000369 |
| 321 |
1 |
0.00037 |
| 326 |
1 |
0.00037 |
| 340 |
1 |
0.00037 |
| 387 |
1 |
0.000369 |
| 400 |
1 |
0.000369 |
| 407 |
1 |
0.000369 |
| 430 |
1 |
0.00037 |
| 435 |
1 |
0.00037 |
| 482 |
1 |
0.00037 |
| 533 |
1 |
0.00037 |
| 541 |
1 |
0.000369 |
| 596 |
1 |
0.00037 |
| 615 |
1 |
0.00037 |
| 642 |
1 |
0.00037 |
| 651 |
1 |
0.000369 |
| 679 |
1 |
0.00037 |
| 689 |
1 |
0.00037 |
| 698 |
1 |
0.00037 |
| 731 |
1 |
0.00037 |
| 751 |
1 |
0.00037 |
| 812 |
1 |
0.00037 |
| 857 |
1 |
0.00037 |
| 880 |
1 |
0.00037 |
| 907 |
1 |
0.00037 |
| 941 |
1 |
0.00037 |
| 943 |
1 |
0.00037 |
| 955 |
1 |
0.00037 |
| 1110 |
1 |
0.00037 |
| 1170 |
1 |
0.00037 |
| 1190 |
1 |
0.00037 |
| 1240 |
1 |
0.00037 |
| 1260 |
1 |
0.00037 |
| 1310 |
1 |
0.00037 |
| 1520 |
1 |
0.00037 |
| 1560 |
1 |
0.00037 |
| 1620 |
1 |
0.00037 |
| 1650 |
1 |
0.00037 |
| 1680 |
1 |
0.00037 |
| 1680 |
1 |
0.00037 |
| 1730 |
1 |
0.00037 |
| 1740 |
1 |
0.00037 |
| 1750 |
1 |
0.00037 |
| 1780 |
1 |
0.00037 |
| 1790 |
1 |
0.00037 |
| 1960 |
1 |
0.00037 |
| 2060 |
1 |
0.00037 |
| 2160 |
1 |
0.00037 |
| 2280 |
1 |
0.00037 |
| 2450 |
1 |
0.00037 |
| 2880 |
1 |
0.00037 |
| 2880 |
1 |
0.00037 |
| 2900 |
1 |
0.00037 |
| 3010 |
1 |
0.00037 |
| 3020 |
1 |
0.00037 |
| 3060 |
1 |
0.00037 |
| 3070 |
1 |
0.00037 |
| 3140 |
1 |
0.00037 |
| 3150 |
1 |
0.00037 |
| 3320 |
1 |
0.00037 |
| 3340 |
1 |
0.00037 |
| 3500 |
1 |
0.00037 |
| 3500 |
1 |
0.00037 |
| 3510 |
1 |
0.00037 |
| 3530 |
1 |
0.00037 |
| 3550 |
1 |
0.00037 |
| 3600 |
1 |
0.00037 |
| 3710 |
1 |
0.00037 |
| 3960 |
1 |
0.00037 |
| 4010 |
1 |
0.00037 |
| 4070 |
1 |
0.00037 |
| 4280 |
1 |
0.00037 |
| 4340 |
1 |
0.00037 |
| 4390 |
1 |
0.00037 |
| 4570 |
1 |
0.00037 |
| 4570 |
1 |
0.00037 |
| 4750 |
1 |
0.000371 |
| 4930 |
1 |
0.00037 |
| 5020 |
1 |
0.00037 |
| 5070 |
1 |
0.00037 |
| 5080 |
1 |
0.00037 |
| 5150 |
1 |
0.00037 |
| 5180 |
1 |
0.00037 |
| 5570 |
1 |
0.00037 |
| 5680 |
1 |
0.00037 |
| 6320 |
1 |
0.00037 |
| 6340 |
1 |
0.00037 |
| 6400 |
1 |
0.00037 |
| 6620 |
1 |
0.000371 |
| 6710 |
1 |
0.00037 |
| 6860 |
1 |
0.00037 |
| 6870 |
1 |
0.00037 |
| 6940 |
1 |
0.000371 |
| 6950 |
1 |
0.00037 |
| 7070 |
1 |
0.00037 |
| 7260 |
1 |
0.00037 |
| 7270 |
1 |
0.00037 |
| 7330 |
1 |
0.00037 |
| 7860 |
1 |
0.00037 |
| 8060 |
1 |
0.00037 |
| 8100 |
1 |
0.00037 |
| 8180 |
1 |
0.00037 |
| 8710 |
1 |
0.00037 |
| 8730 |
1 |
0.00037 |
| 9310 |
1 |
0.00037 |
| 9320 |
1 |
0.00037 |
| 9560 |
1 |
0.00037 |
| 9900 |
1 |
0.00037 |
| 10000 |
1 |
0.00037 |
| 10100 |
1 |
0.00037 |
| 10600 |
1 |
0.00037 |
| 10600 |
1 |
0.00037 |
| 10700 |
1 |
0.00037 |
| 10700 |
1 |
0.00037 |
| 10800 |
1 |
0.00037 |
| 11000 |
1 |
0.00037 |
| 11100 |
1 |
0.00037 |
| 12100 |
1 |
0.00037 |
| 12900 |
1 |
0.00037 |
| 13000 |
1 |
0.00037 |
| 13000 |
1 |
0.00037 |
| 13900 |
1 |
0.00037 |
| 14100 |
1 |
0.00037 |
| 15100 |
1 |
0.00037 |
| 15200 |
1 |
0.00037 |
| 15500 |
1 |
0.00037 |
| 15700 |
1 |
0.00037 |
| 15700 |
1 |
0.00037 |
| 15800 |
1 |
0.00037 |
| 15800 |
1 |
0.00037 |
| 16400 |
1 |
0.00037 |
| 16500 |
1 |
0.00037 |
| 16600 |
1 |
0.00037 |
| 16800 |
1 |
0.00037 |
| 16800 |
1 |
0.00037 |
| 16900 |
1 |
0.00037 |
| 17500 |
1 |
0.00037 |
| 17500 |
1 |
0.00037 |
| 17500 |
1 |
0.00037 |
| 17600 |
1 |
0.00037 |
| 17600 |
1 |
0.00037 |
| 17600 |
1 |
0.00037 |
| 18400 |
1 |
0.00037 |
| 19300 |
1 |
0.00037 |
| 19500 |
1 |
0.00037 |
| 20100 |
1 |
0.00037 |
| 20400 |
1 |
0.00037 |
| 20600 |
1 |
0.00037 |
| 20700 |
1 |
0.00037 |
| 20900 |
1 |
0.00037 |
| 21400 |
1 |
0.00037 |
| 21500 |
1 |
0.00037 |
| 22600 |
1 |
0.00037 |
| 22700 |
1 |
0.00037 |
| 22800 |
1 |
0.00037 |
| 22800 |
1 |
0.00037 |
| 23000 |
1 |
0.00037 |
| 23500 |
1 |
0.00037 |
| 23600 |
1 |
0.00037 |
| 23800 |
1 |
0.00037 |
| 23900 |
1 |
0.00037 |
| 24000 |
1 |
0.00037 |
| 24400 |
1 |
0.00037 |
| 24600 |
1 |
0.00037 |
| 24800 |
1 |
0.00037 |
| 25000 |
1 |
0.00037 |
| 25100 |
1 |
0.00037 |
| 25300 |
1 |
0.00037 |
| 25400 |
1 |
0.00037 |
| 25600 |
1 |
0.00037 |
| 25700 |
1 |
0.00037 |
|
CL1 ----> CL14

No. of shared pairs: :377
CL1 ----> CL7

No. of shared pairs: :349
CL1 ----> CL46
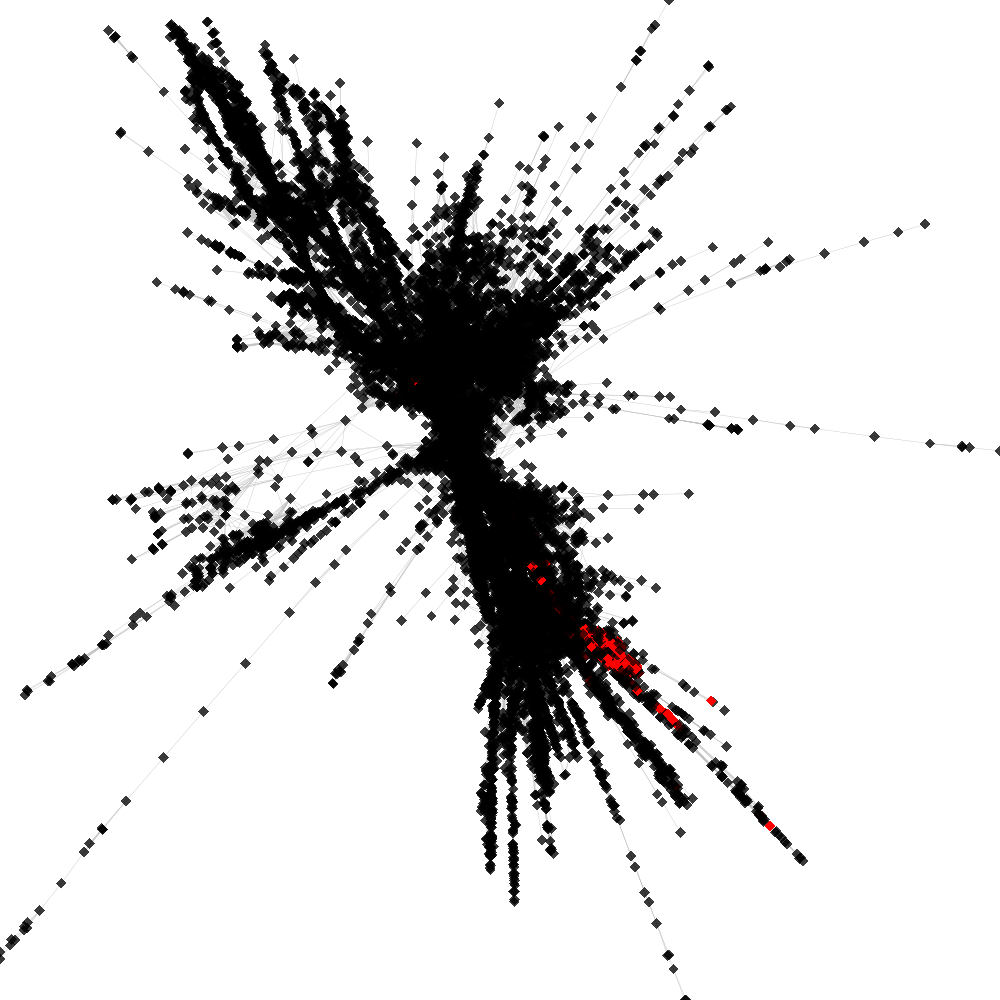
No. of shared pairs: :330
CL1 ----> CL45
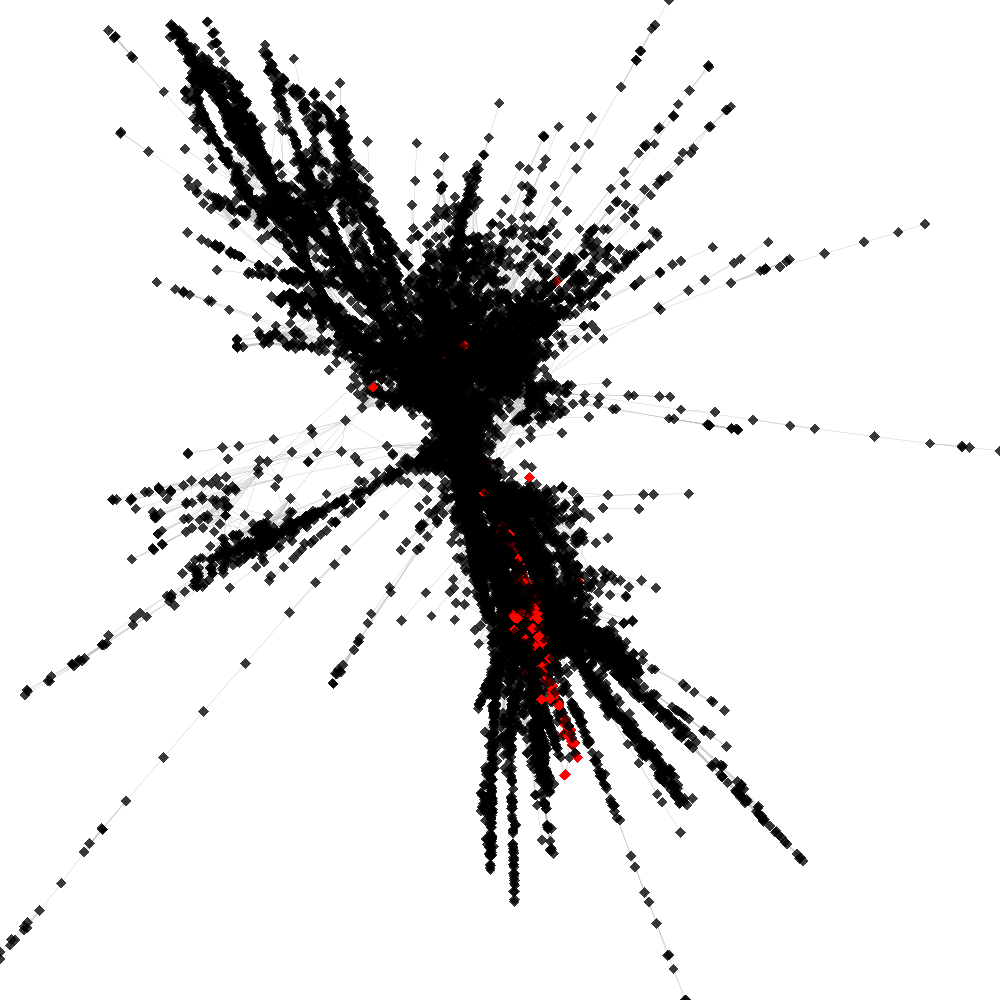
No. of shared pairs: :212
CL1 ----> CL62
No. of shared pairs: :181
CL1 ----> CL28

No. of shared pairs: :178
CL1 ----> CL51

No. of shared pairs: :154
CL1 ----> CL34

No. of shared pairs: :151
CL1 ----> CL65

No. of shared pairs: :149
CL1 ----> CL75

No. of shared pairs: :147
CL1 ----> CL54

No. of shared pairs: :139
CL1 ----> CL22

No. of shared pairs: :118
CL1 ----> CL66

No. of shared pairs: :111
CL1 ----> CL87

No. of shared pairs: :111
CL1 ----> CL10
No. of shared pairs: :100
CL1 ----> CL67

No. of shared pairs: :85
CL1 ----> CL71

No. of shared pairs: :83
CL1 ----> CL37

No. of shared pairs: :70
CL1 ----> CL15

No. of shared pairs: :58
CL1 ----> CL42
No. of shared pairs: :49
CL1 ----> CL105

No. of shared pairs: :37
CL1 ----> CL89

No. of shared pairs: :36
CL1 ----> CL2
No. of shared pairs: :28
CL1 ----> CL12

No. of shared pairs: :28
CL1 ----> CL90
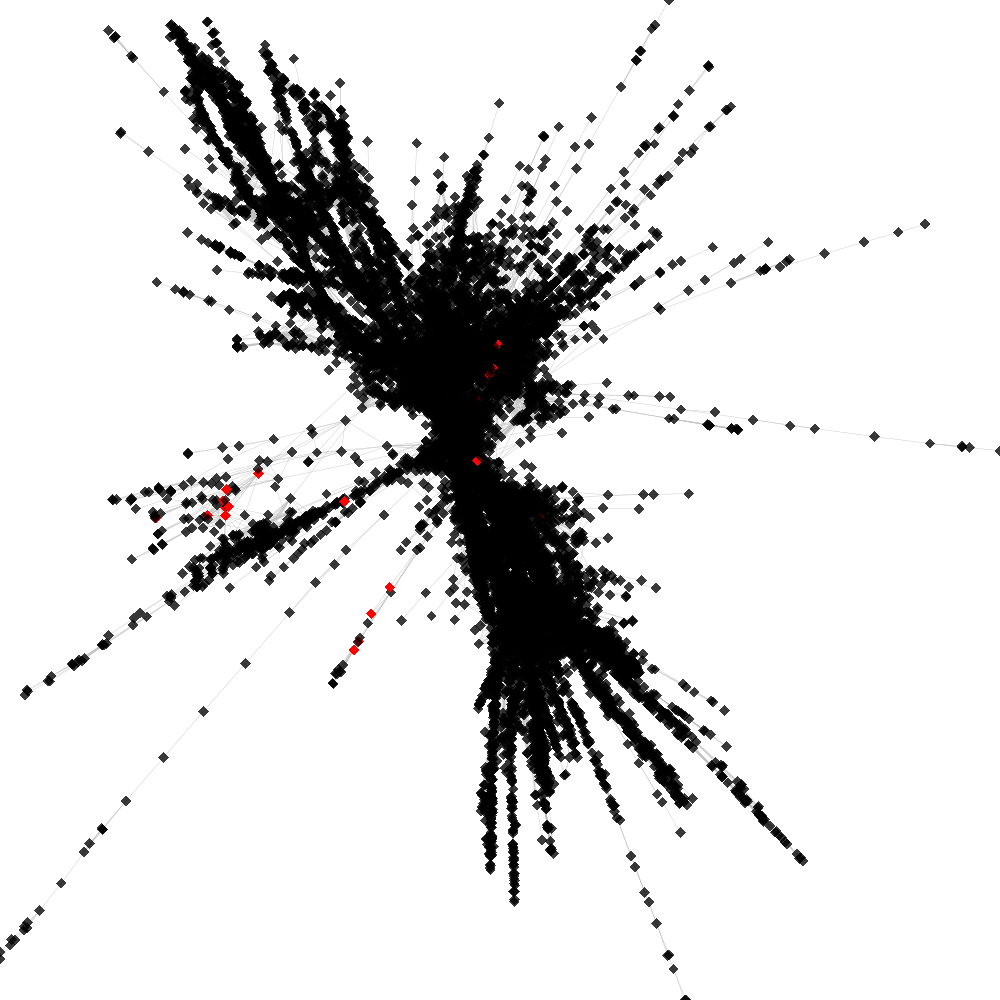
No. of shared pairs: :28
CL1 ----> CL200

No. of shared pairs: :24
CL1 ----> CL3
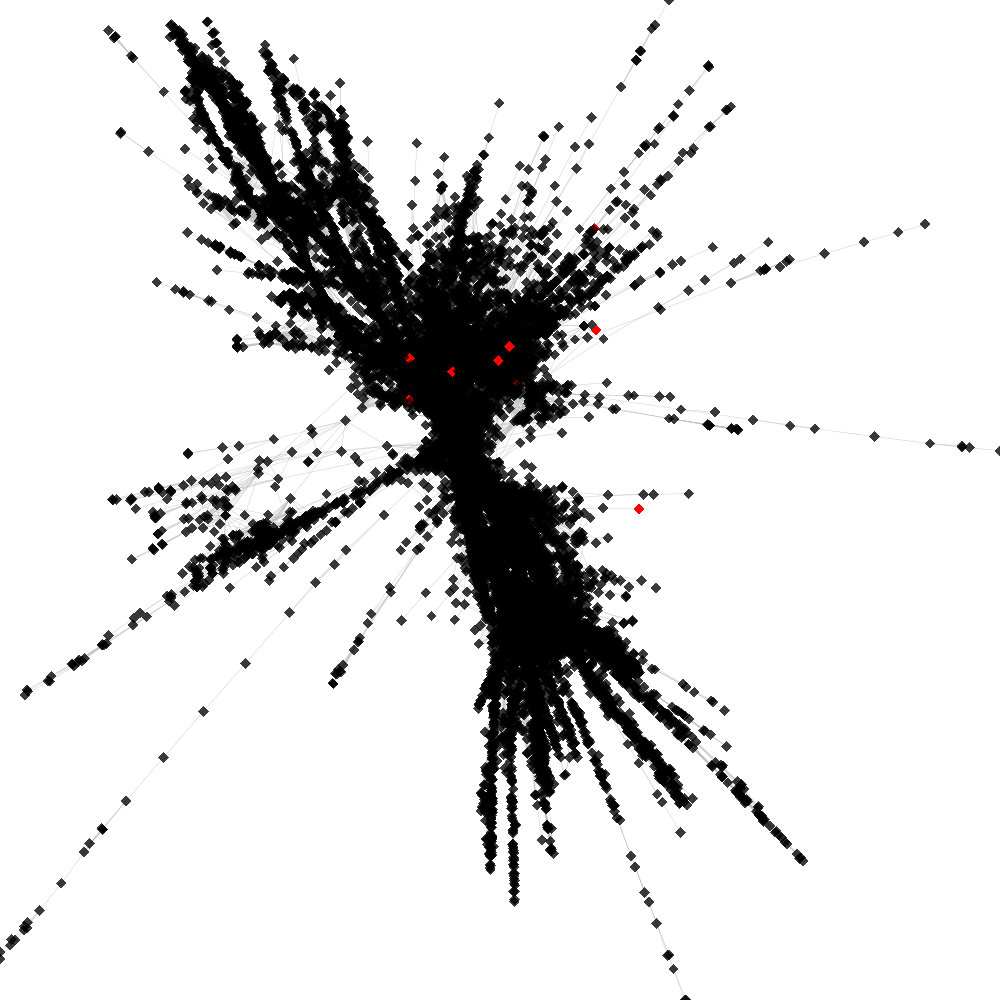
No. of shared pairs: :23
CL1 ----> CL127

No. of shared pairs: :23
CL1 ----> CL5

No. of shared pairs: :21
CL1 ----> CL13

No. of shared pairs: :20
CL1 ----> CL17

No. of shared pairs: :18
CL1 ----> CL23

No. of shared pairs: :17
CL1 ----> CL102

No. of shared pairs: :17
CL1 ----> CL6

No. of shared pairs: :15
CL1 ----> CL16
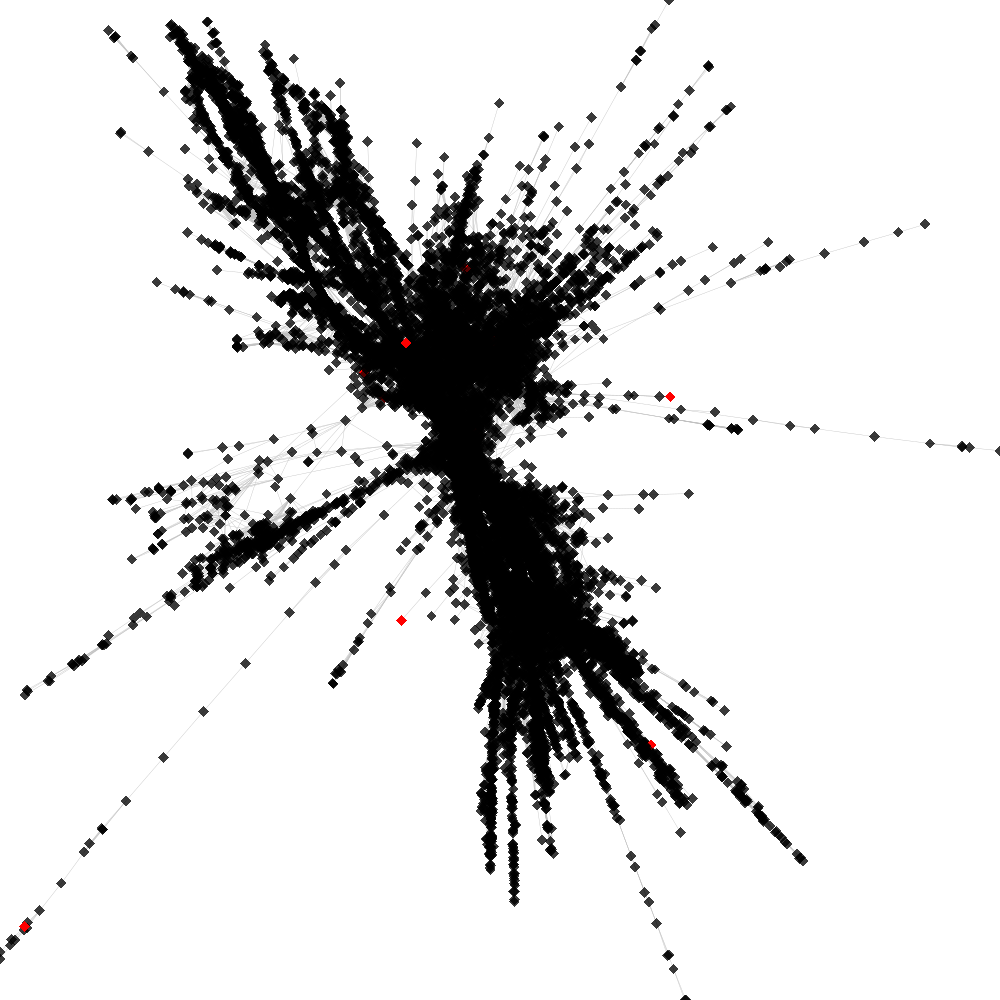
No. of shared pairs: :15
CL1 ----> CL8

No. of shared pairs: :13
CL1 ----> CL21

No. of shared pairs: :12
CL1 ----> CL24

No. of shared pairs: :12
CL1 ----> CL25

No. of shared pairs: :12
CL1 ----> CL36

No. of shared pairs: :12
CL1 ----> CL218

No. of shared pairs: :12


