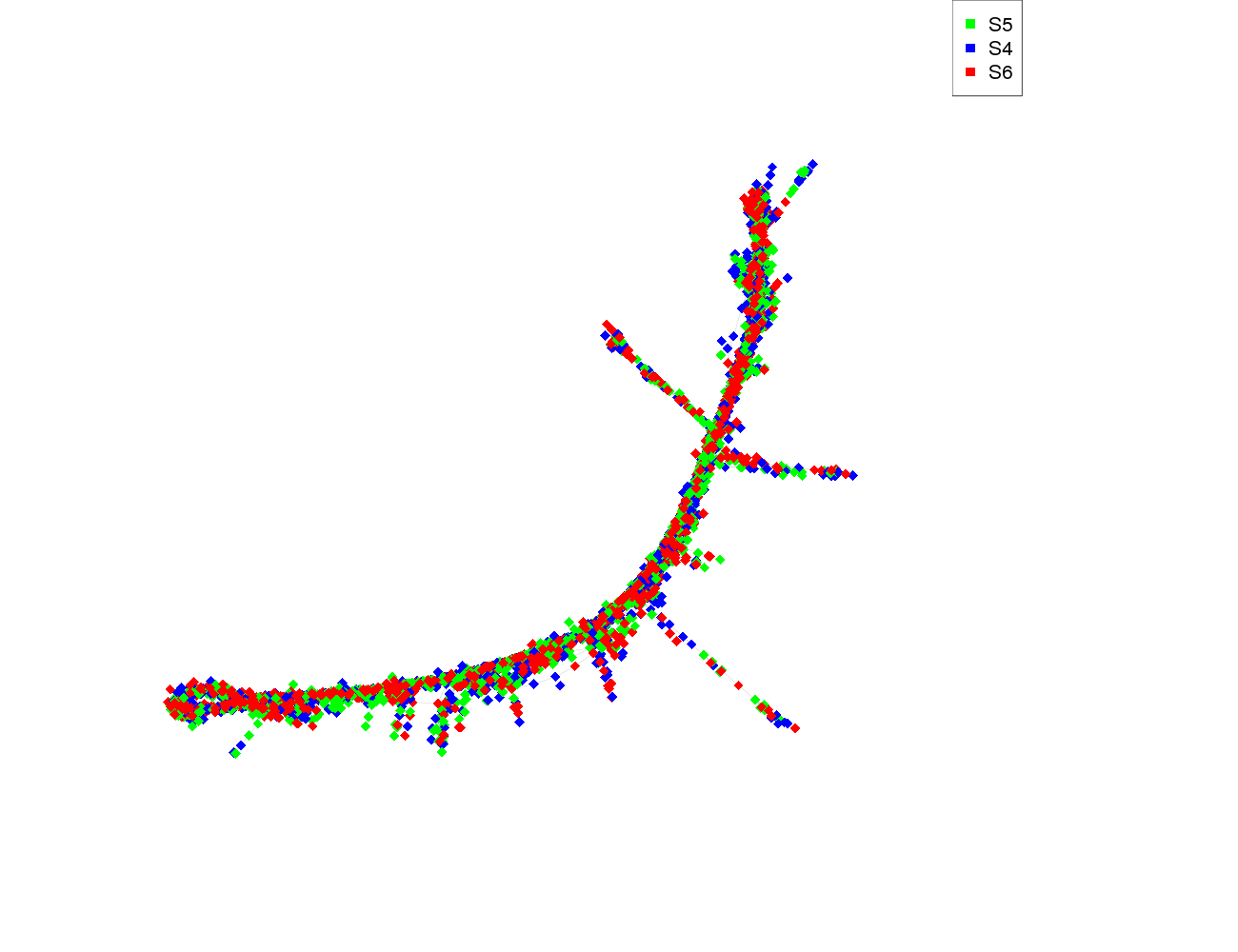
| size | 3522 |
| size_real | 3522 |
| ecount | 59381 |
| supercluster | 6 |
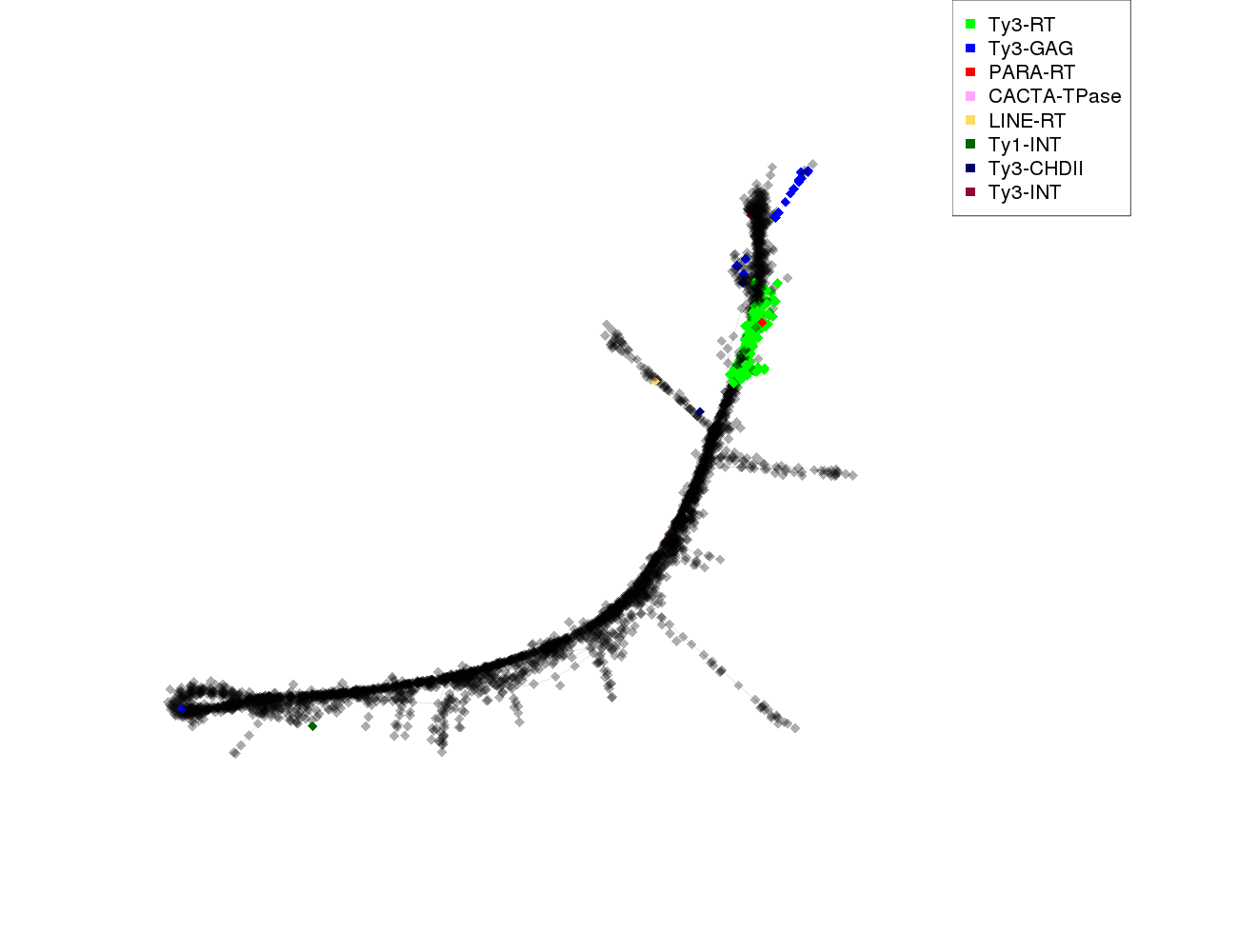
| annotations_summary | 5.65% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RT 0.45% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-GAG 0.14% Class_I/LTR/Ty3_gypsy/chromovirus/Chlamyvir:Ty3-RT 0.09% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-RT 0.09% Class_I/LTR/Ty3_gypsy/chromovirus/Tcn1:Ty3-RT 0.09% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-RT 0.06% Class_I/LTR/Ty3_gypsy/chromovirus/Galadriel:Ty3-RT 0.06% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-RT 0.06% Class_I/pararetrovirus:PARA-RT 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-CHDII 0.03% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-GAG 0.03% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatII:Ty3-INT 0.03% Class_I/LTR/Ty1_copia/Ivana:Ty1-INT 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-GAG 0.03% Class_II/Subclass_1/TIR/EnSpm_CACTA:CACTA-TPase 0.03% Class_I/LINE:LINE-RT |
| pair_completeness | 0.795107033639144 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.000851788756388416 |
| satellite_probability | 2.63652108321905e-23 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S5 | 1200 |
| S4 | 1240 |
| S6 | 1090 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 6590 | 7100 | 6150 |
| S4 | 7100 | 7410 | 6630 |
| S6 | 6150 | 6630 | 5630 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 0.994 | 1.010 | 1.000 |
| S4 | 1.010 | 0.985 | 1.010 |
| S6 | 1.000 | 1.010 | 0.987 |
protein domains:
|
|
||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|