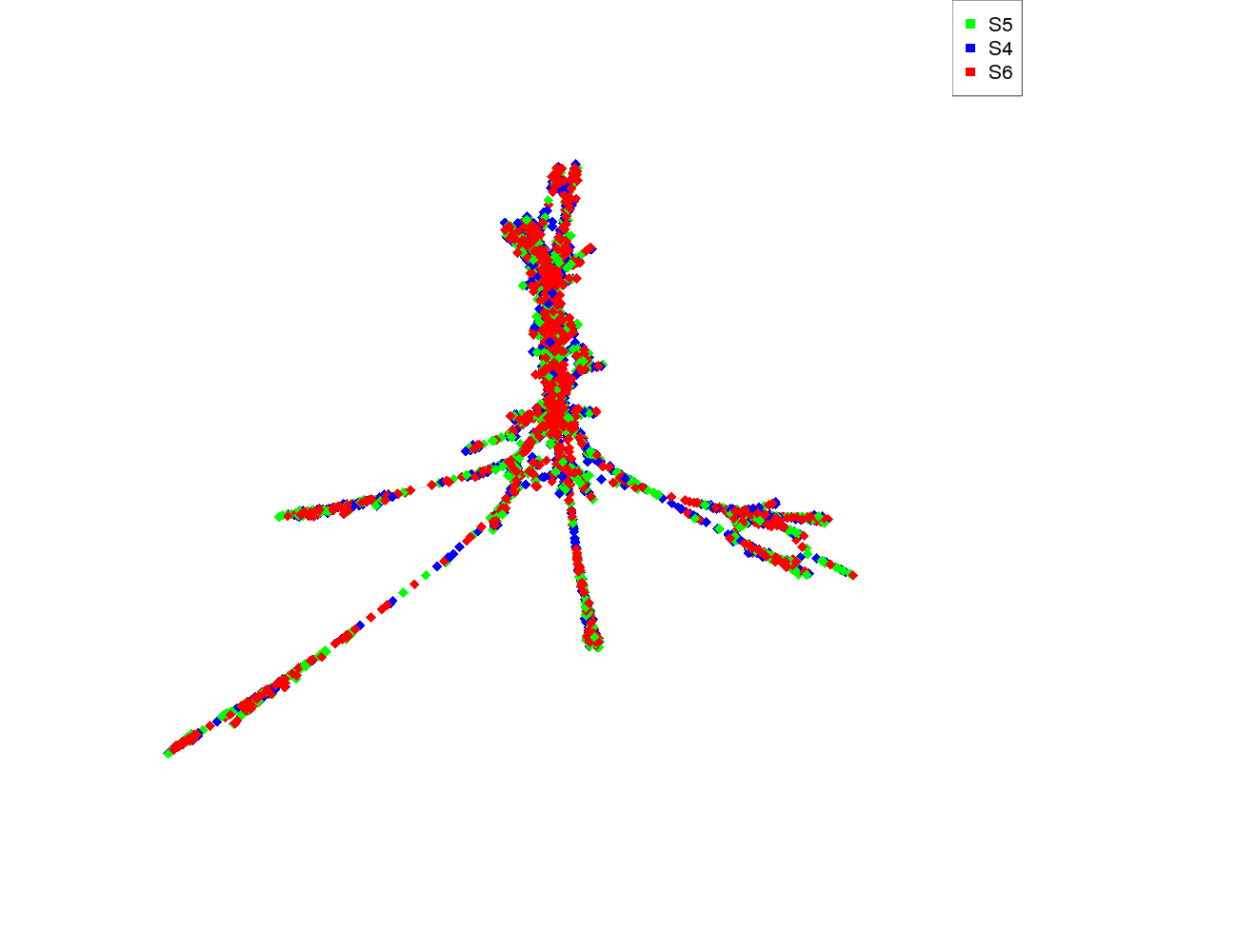
| size | 4106 |
| size_real | 4106 |
| ecount | 22436 |
| supercluster | 13 |
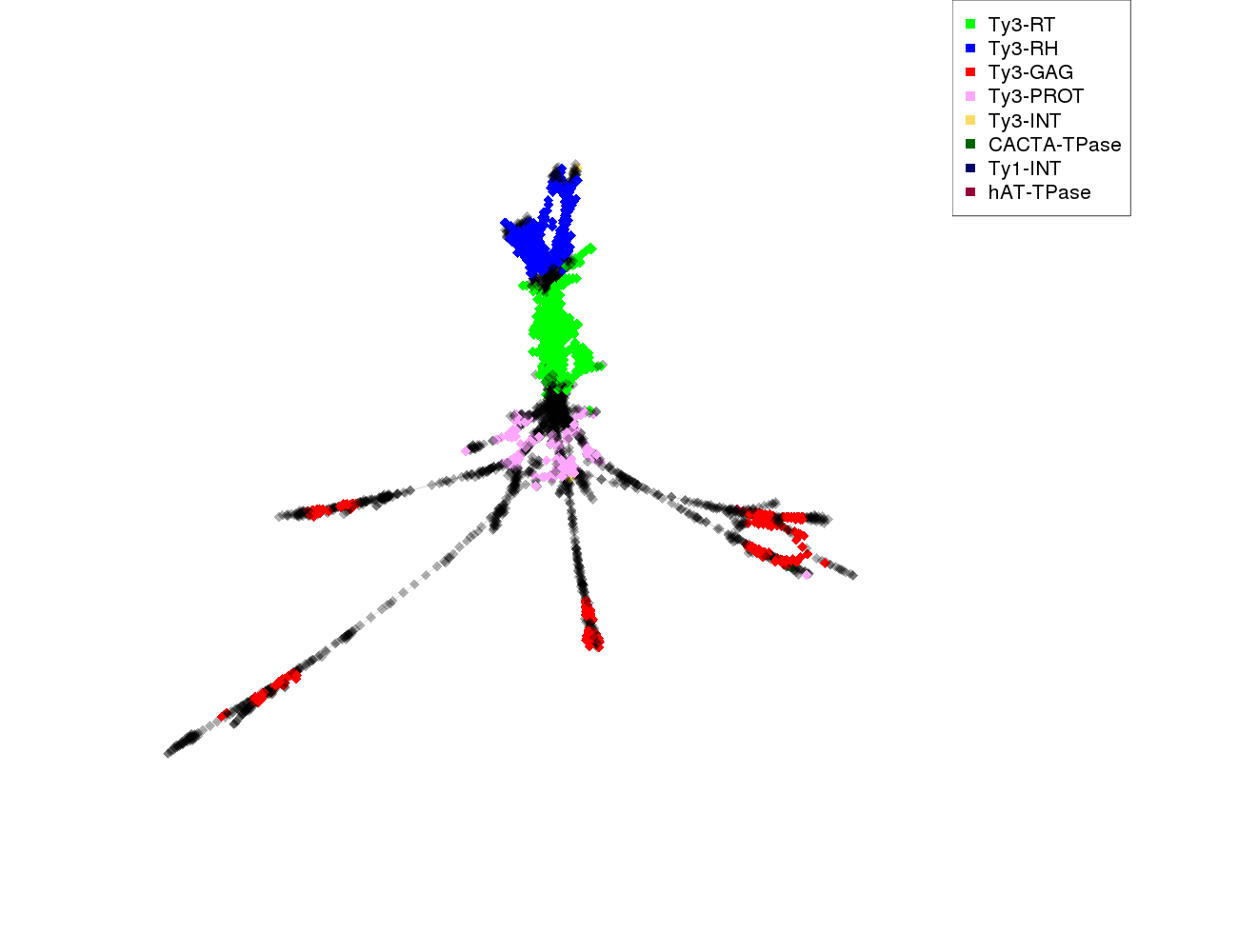
| annotations_summary | 17.54% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RH 16.39% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RT 7.48% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-GAG 6.02% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-PROT 1.32% Class_I/LTR/Ty3_gypsy/chromovirus/Chlamyvir:Ty3-RT 1.19% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-GAG 0.71% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-RT 0.22% Class_I/LTR/Ty3_gypsy/chromovirus/Galadriel:Ty3-RT 0.10% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-GAG 0.10% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-PROT 0.07% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-RH 0.07% Class_I/LTR/Ty3_gypsy/chromovirus/chromo-unclass:Ty3-RT 0.07% Class_I/LTR/Ty3_gypsy/chromovirus/Tcn1:Ty3-RT 0.07% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-RH 0.05% Class_I/LTR/Ty3_gypsy/chromovirus/Galadriel:Ty3-RH 0.05% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-RH 0.05% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-RT 0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-RH 0.02% Class_I/LTR/Ty3_gypsy/chromovirus/Chlamyvir:Ty3-PROT 0.02% Class_II/Subclass_1/TIR/EnSpm_CACTA:CACTA-TPase 0.02% Class_II/Subclass_1/TIR/hAT:hAT-TPase 0.02% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-INT 0.02% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-INT 0.02% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-INT |
| pair_completeness | 0.682786885245902 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.000243546030199708 |
| satellite_probability | 2.29499556110385e-22 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S5 | 1380 |
| S4 | 1380 |
| S6 | 1350 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 2610 | 2510 | 2530 |
| S4 | 2510 | 2410 | 2440 |
| S6 | 2530 | 2440 | 2460 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 1.000 | 1.000 | 0.999 |
| S4 | 1.000 | 0.999 | 1.000 |
| S6 | 0.999 | 1.000 | 1.000 |
protein domains:
|
|
||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|