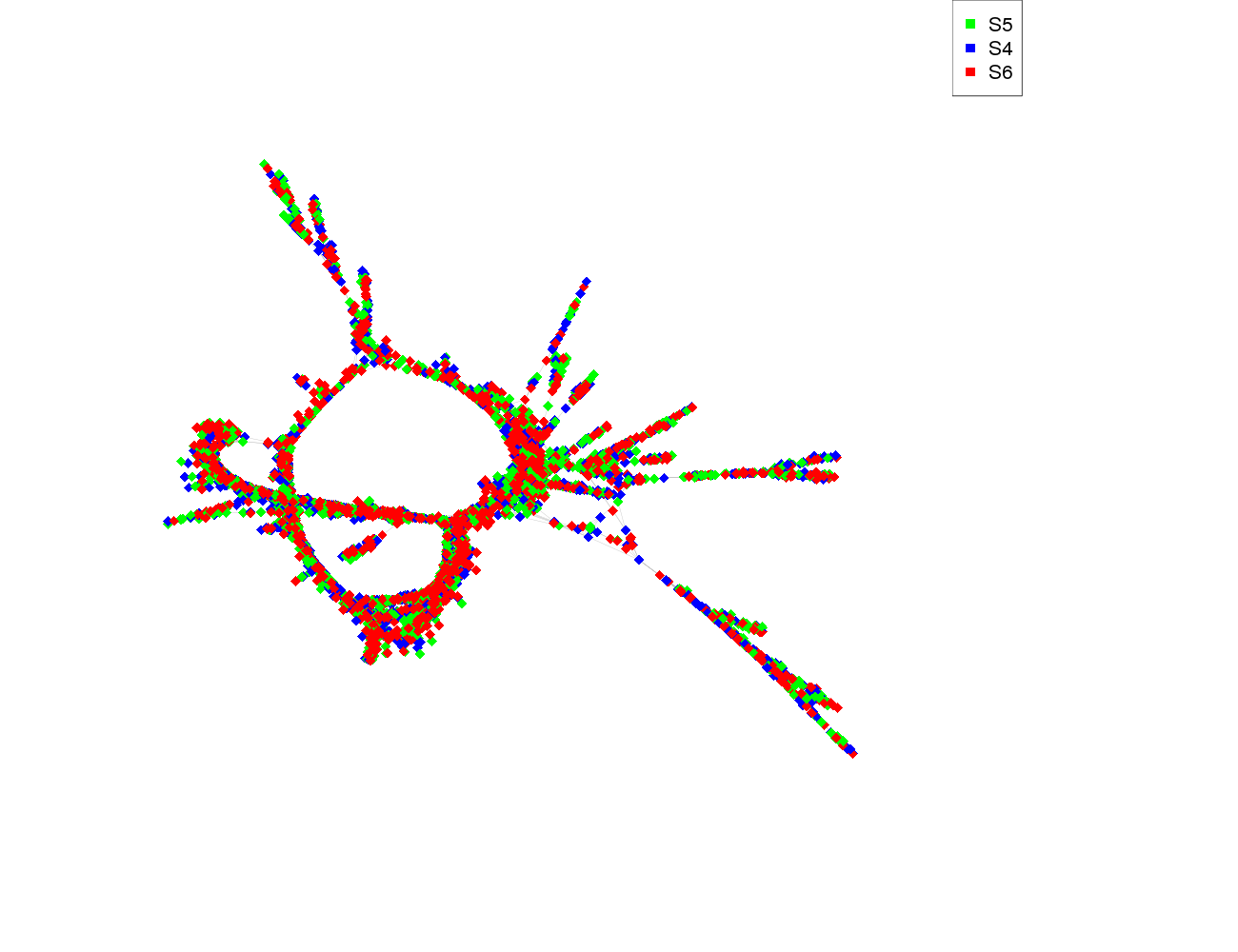
| size | 5123 |
| size_real | 5123 |
| ecount | 53859 |
| supercluster | 10 |
| annotations_summary | 7.28% Class_I/LTR/Ty1_copia/SIRE:Ty1-GAG 0.29% Class_I/LTR/Ty1_copia/TAR:Ty1-INT 0.21% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-GAG 0.16% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-GAG 0.06% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-INT 0.04% Class_II/Subclass_1/TIR/hAT:hAT-TPase 0.04% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RH 0.02% organelle/mitochondria 0.02% Class_I/LTR/Ty3_gypsy/chromovirus/Chlamyvir:Ty3-INT 0.02% Class_II/Subclass_1/TIR/Tc1_Mariner:Mariner-TPase 0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-RT 0.02% Class_I/LTR/Ty1_copia/Ale:Ty1-GAG 0.02% Class_I/LTR/Ty1_copia/Ivana:Ty1-GAG 0.02% Class_I/LTR/Ty1_copia/SIRE:Ty1-INT 0.02% Class_II/Subclass_1/TIR/EnSpm_CACTA:CACTA-TPase 0.02% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-INT 0.02% Class_I/pararetrovirus:PARA-RT |
| pair_completeness | 0.672543258243552 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.597501463985946 |
| satellite_probability | 0.000985291075113611 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S5 | 1750 |
| S4 | 1660 |
| S6 | 1710 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 6260 | 6010 | 5990 |
| S4 | 6010 | 6010 | 5920 |
| S6 | 5990 | 5920 | 5740 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 1.010 | 0.988 | 1.000 |
| S4 | 0.988 | 1.010 | 1.010 |
| S6 | 1.000 | 1.010 | 0.992 |
protein domains:
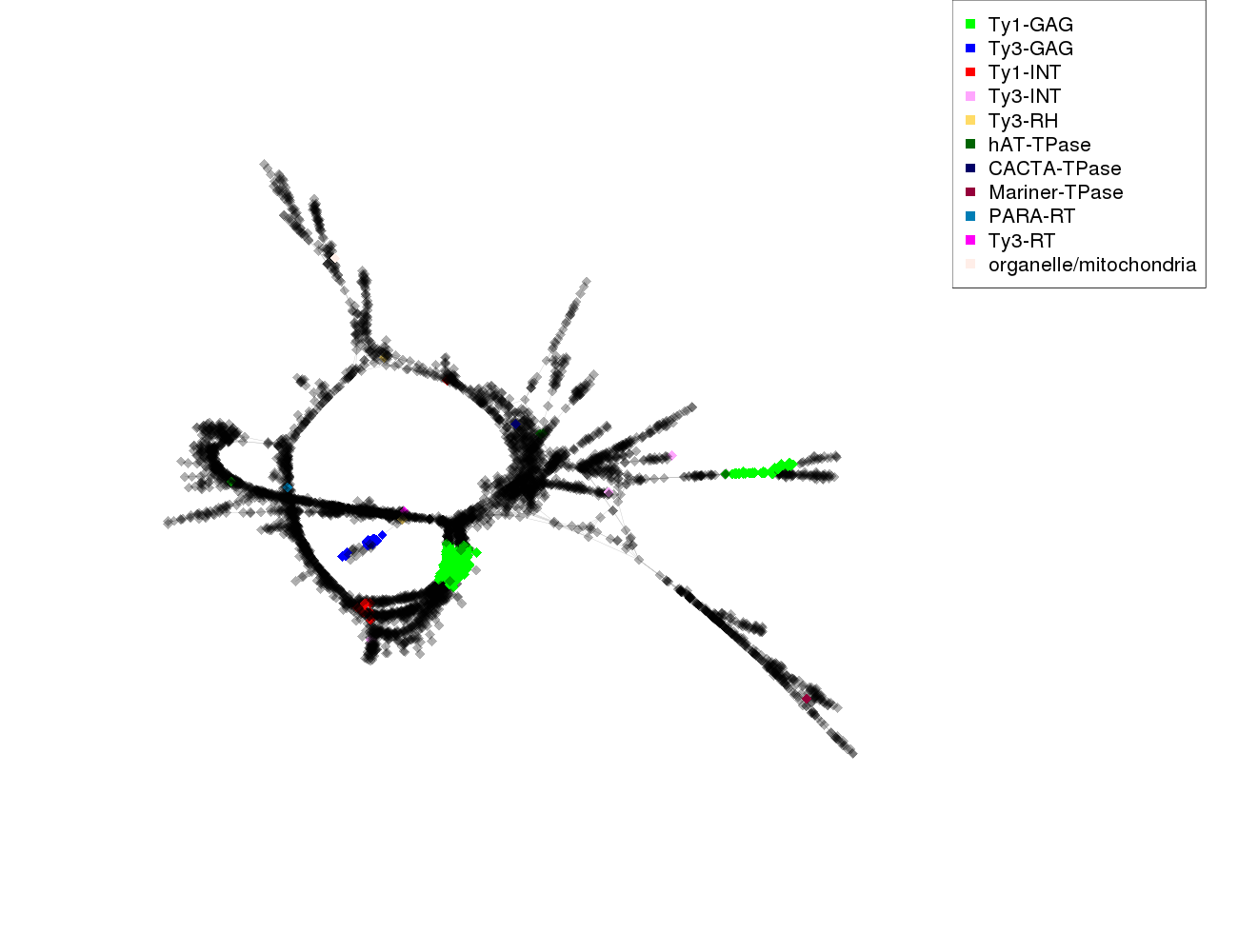
|
|
||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|