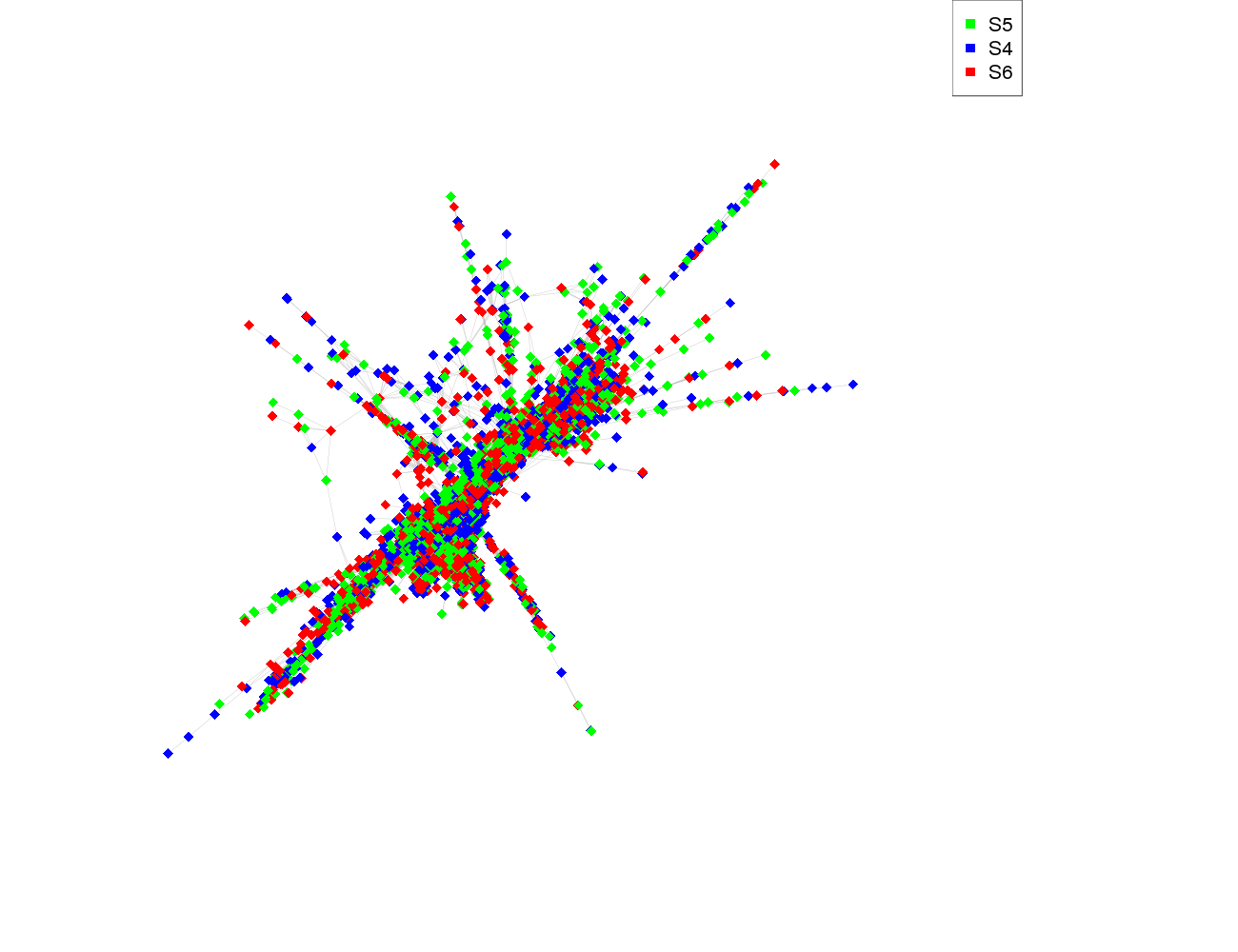
| size | 5864 |
| size_real | 5864 |
| ecount | 538556 |
| supercluster | 1 |
| annotations_summary | 0.03% organelle/plastid 0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIII:Ty3-RH 0.02% Class_II/Subclass_1/TIR/hAT:hAT-TPase |
| pair_completeness | 0.483805668016194 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.095668485675307 |
| satellite_probability | 1.65284900887401e-15 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

| Species | Read count |
|---|---|
| S5 | 1960 |
| S4 | 1930 |
| S6 | 1980 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 57100 | 57600 | 60400 |
| S4 | 57600 | 58900 | 61300 |
| S6 | 60400 | 61300 | 64000 |
| S5 | S4 | S6 | |
|---|---|---|---|
| S5 | 1.000 | 0.997 | 1 |
| S4 | 0.997 | 1.000 | 1 |
| S6 | 1.000 | 1.000 | 1 |
protein domains:
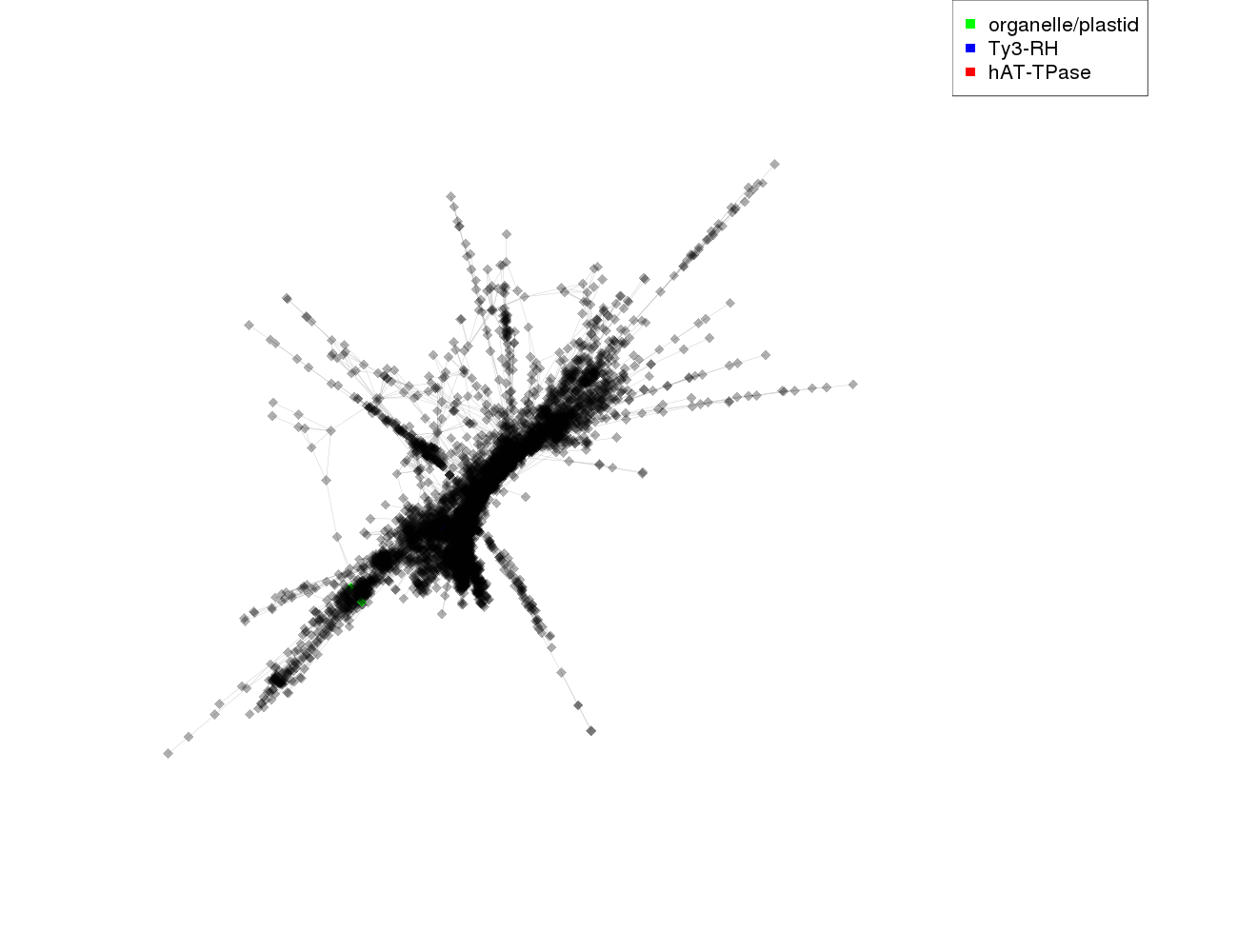
|
|
||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|