| size | 2989 |
| size_real | 2989 |
| ecount | 12551 |
| supercluster | 27 |
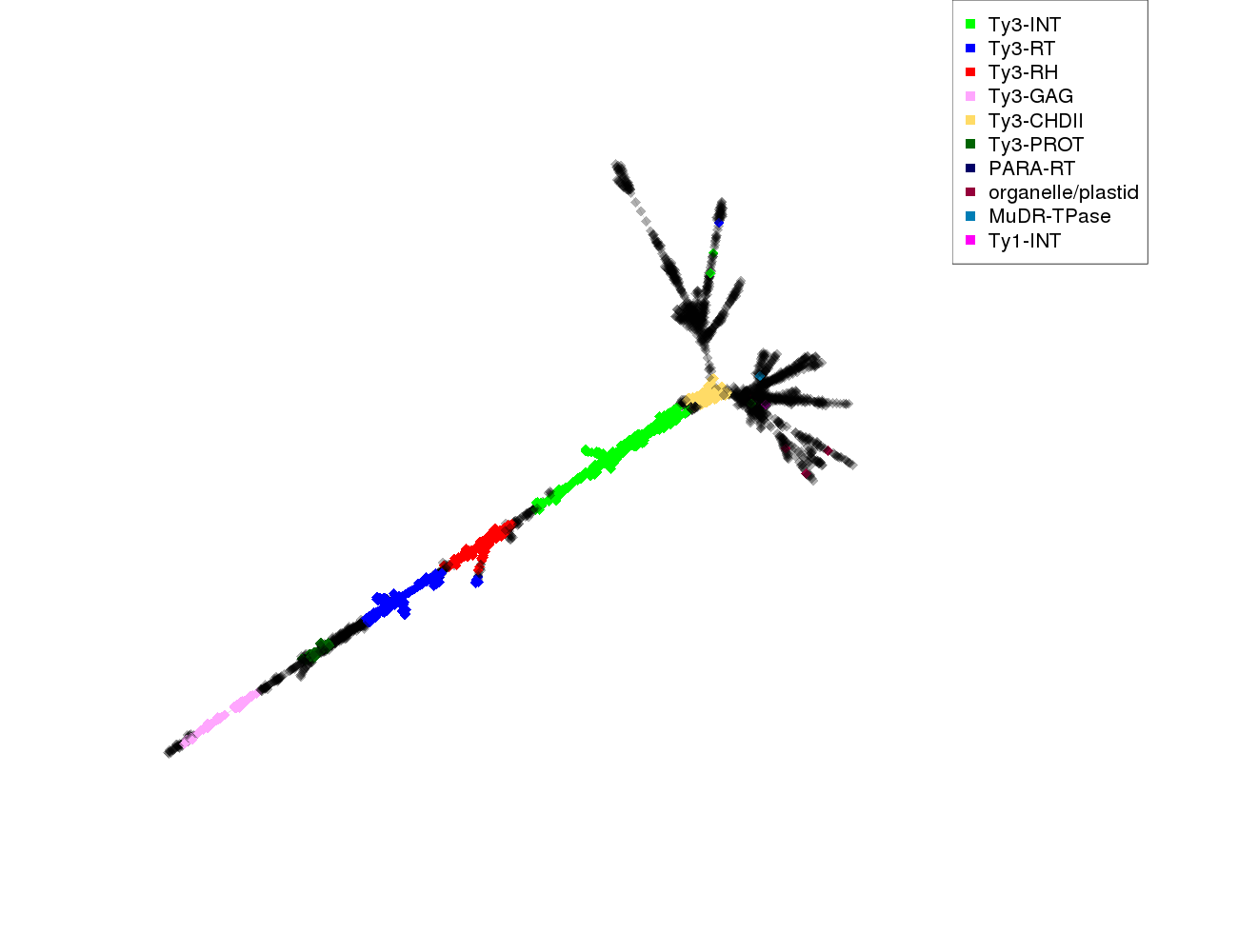
| annotations_summary | 14.85% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-INT 7.46% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RT 6.96% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-RH 2.81% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-GAG 2.38% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-CHDII 1.67% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-PROT 0.54% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-RT 0.40% Class_I/LTR/Ty3_gypsy/chromovirus/Chlamyvir:Ty3-RT 0.27% Class_I/pararetrovirus:PARA-RT 0.23% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-CHDII 0.17% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-RH 0.13% organelle/plastid 0.10% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-GAG 0.10% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-PROT 0.07% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-RT 0.07% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-INT 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-RH 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/Tcn1:Ty3-INT 0.03% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-RT 0.03% Class_II/Subclass_1/TIR/MuDR_Mutator:MuDR-TPase 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/Galadriel:Ty3-RH 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-RH 0.03% Class_I/LTR/Ty1_copia/Bianca:Ty1-INT 0.03% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-INT |
| pair_completeness | 0.600964113551152 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.000334560053529609 |
| satellite_probability | 7.63705200701412e-22 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |

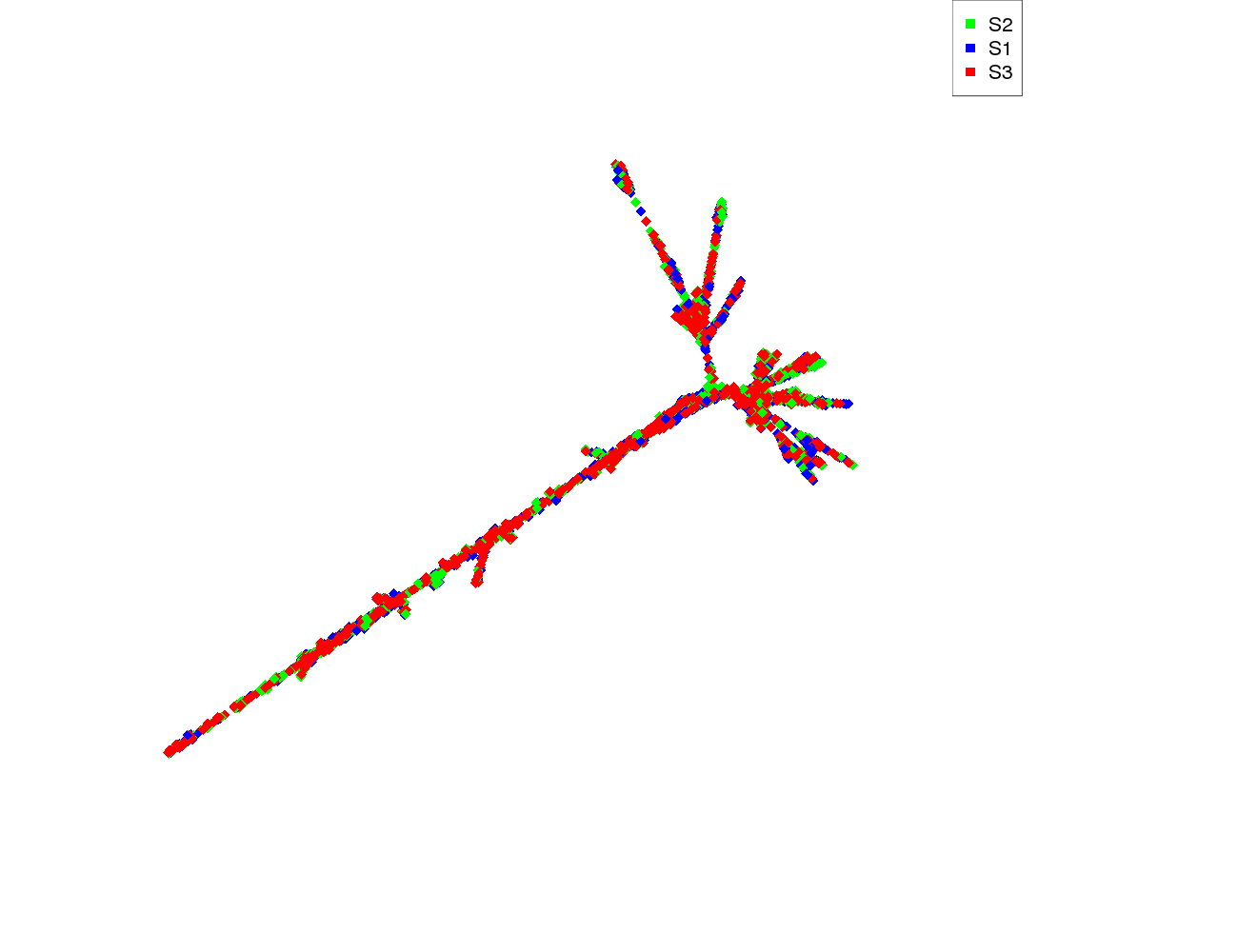
| Species | Read count |
|---|---|
| S2 | 934 |
| S1 | 999 |
| S3 | 1060 |
| S2 | S1 | S3 | |
|---|---|---|---|
| S2 | 1240 | 1320 | 1320 |
| S1 | 1320 | 1580 | 1490 |
| S3 | 1320 | 1490 | 1490 |
| S2 | S1 | S3 | |
|---|---|---|---|
| S2 | 1.030 | 0.976 | 0.994 |
| S1 | 0.976 | 1.030 | 0.991 |
| S3 | 0.994 | 0.991 | 1.010 |
protein domains:
|
|
||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|